Introduction
The espl1 (extra spindle pole bodies like 1) gene encodes separase protein, which is a site-specific peptidase of the CD group of cysteine proteinases.1 Apart from separase, CD clan group also includes other protein families like caspase-1 (C14), clostripain (C11), gingipain R (C25), legumain (C13), and separin (C50).2 Separase is a key molecule for separating mitotic sister chromatids during mitosis and meiosis. For the first time, separase protein was found to regulate spindle formation in fungi.3 In the present study, the functions of separating enzymes are mainly reflected in four aspects: Chromosome cycle, Centrosome cycle, DNA damage repair, and Membrane trafficking.4,5 Researchers have done some research on the diseases caused by separase. Separase is an oncogene, which is overexpressed in many human cancers of the breast, bone, brain, and prostate.6–8 During meiosis, espl1 for encoding separase protein is a mitotic key player in chromosomal segregation and germ cell development. In animals, the germline is designated as a primordial germ cell (PGCs) in the early stages of embryogenesis. PGCs critically depend on the inhibitory phosphorylation of separase to prevent premature separation of sister chromatids and, hence, progeny with abnormal chromosome numbers.9,10 Probably, espl1 plays an important role in the development of vertebrate PGCs and gametogenesis. In teleosts, the activities of gonads are primarily regulated by the hypothalamic-pituitary-gonadal (HPG) axis,11 and LHRH-A2 (luteinizing hormone-releasing hormone, an analog of GnRH) is important endogenous hormone analogs of the reproductive axis that enable maturation of the gonads.12 As such, LHRH-A2 is widely used to study the expression of genes of gonadal development in fish.
The loach (Misgurnus anguillicaudatus) is a small benthic species belonging to the family Cobitidae. This species is widely distributed in midstream and downstream regions in Korea, China, and Taiwan. Loach is a polyploid fish, in addition to diploids in natural waters, there are also triploid, tetraploid, and hexaploid, and their chromosome complement is very complicated. Therefore, loach is an ideal biological model for studying the function of the espl1 gene. So far, the espl1 gene for encoding separase protein has not been studied in fish. Therefore, the role of espl1 in the gonadal development of loach was discussed in this paper. In this study, the full length of the espl1 gene of loach was first cloned and characterized by using bioinformatics methods and analysis of its specific expression of early embryonic development by whole-mount in situ hybridization. At the same time, to primarily study the distribution of espl1 in loach, multi-tissue Real-time quantitative PCR (qPCR) was applied to determine its tissue-specific expression in various organ tissues from adult loach, and its Gonads Specific Expression by qPCR and LHRH-A2. The results suggested that the loach espl1 gene is highly conserved in vertebrates and specifically expressed in the gonads of adult loach. The espl1 gene plays an important role in loach during gonad development.
Materials and Methods
Fish
The fish used in the experiment were wild diploid adult loaches from waters near Ezhou city of Hubei province in China. Diploid loaches were chosen by a ploidy analyzer (Partec, Germany) and divided into three groups. The first group was used for cloning the espl1 gene and investigating tissue expressions of the espl1 gene. The second group was used for reproduction to study the expression of espl1 gene at different stages of embryonic development by whole-mount in situ hybridization. The third group of fish was used to study espl1 gene expression levels by injecting different concentrations of LHRH-A2.
Cloning the full-length cDNAs of espl1
Total RNA was isolated from adult loaches from different tissues (heart, kidney, liver, spleen, intestine, muscle, skin, testis, ovary, brain, gill) using TRIzol Reagent (Takara, Japan) according to the manufacturer’s instructions. The cDNA was synthesized using a Yeomen’s reverse transcription kit. The cDNA was kept at -20 ℃ for long-term storage.
The primers listed in (Table 1) were used for partial sequences of espl1 gene amplification, and Primer sequences are designed by Primer 5.0 software. The full-length cDNAs of espl1 sequence includes four sequences: sequence-1, sequence-2, sequence-3, sequence-4. Parameters of the PCR program were listed as follows: sequence-1 (414 bp): initial denaturation at 98 ℃ for 3 min, then 35 cycles of 10 s at 98 ℃, 20 s at 60 ℃, 10 s at 72 ℃, and final extension at 72 ℃ for 5 min. sequence-2 (1516 bp): initial denaturation at 94 ℃ for 5 min, then 35 cycles of 30 s at 94 ℃, 30 s at 60 ℃, 91 s at 72 ℃, and final extension at 72 ℃ for 5 min. sequence-3 (1953 bp): initial denaturation at 94 ℃ for 5 min, then 35 cycles of 30 s at 94 ℃, 30 s at 55 ℃, 120 s at 72 ℃, and final extension at 72 ℃ for 5 min, and sequence-4 (1027 bp): initial denaturation at 94 ℃ for 5 min, then 35 cycles of 30 s at 94 ℃, 30 s at 60 ℃, 61 s at 72 ℃, and final extension at 72 ℃ for 5 min. The PCR products were collected with 2% agarose (Sangon, China) and then purified with a TaKaRa Agarose Gel DNA Purification Kit Ver.2.0 (TaKaRa). After purification, the DNA fragments were ligated into the PMD19-T and transformed into competent Escherichia coli DH5α cells. Randomly selected clones were sequenced (Invitrogen).
Sample collections and Real-time quantitative PCR (qPCR)
To investigate the expression levels of the espl1 gene in different tissues, 14 tissues (ovary, eye, fin, heart, skin, muscle, testis, liver, brain, beard, intestine, gill, spleen, and kidney) were dissected and stored at -80 ℃, and Selecting GAPDH and β-actin as internal controls for cDNA standards. Parameters of the qPCR program were listed as fellow: 94 ℃ for 15 s, 60 s at 60 ℃, and 10 s at 72 ℃, for extension, repeating for 40 cycles, and each tissue acted as a biological repeat (n = 3). The qPCR primers are listed in Table 1.
Whole-mount in situ hybridization
To study the expression of the espl1 gene in early loach development, the antisense probe for espl1 was synthesized using T7 RNA polymerase (Roche) with an EcoRI-linearized plasmid as a template. The probe was an in vitro transcription product of a 513-773 fragment of the espl1 cDNA.
The whole-mount and gonads in situ hybridization were performed as described previously (see http://zfin.org/zf_info/zfbook/chapt9/9.82.html).13
Sequence analysis and construction of phylogenetic tree
The amino acid sequences of espl1 from different species were obtained by NCBI Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi). BioEdit software conducted multiple alignments of the deduced amino acid sequences. The phylogenetic trees of espl1 amino acid sequences were constructed by MEGA5.0 using the neighbor-joining method (Arizona State University, USA), and the reliability of the branching was tested using bootstrap resampling (1000 pseudo-replicates).
Effects of LHRH-A2 on espl1 expression in gonads of adult loach
The experiment in this study included three treatments with three replicates, and each treatment includes three mature males and females loach. each group containing three replicates of fish: a control group (injected with saline), a low LHRH-A2 group (injected with 0.05 μg/g bodyweight), and a high LHRH-A2 group (injected with 0.2 μg/g bodyweight). The injections were performed on day 1 and day 4, and the gonad collection was performed on day 5. All the samples were quickly dissected, snap frozen, stored in liquid nitrogen, and then processed for RNA extraction, and different groups of mature gonads were selected for tissue section.
Statistical analysis
The qPCR data were given in the form of relative mRNA and shown as mean ± SE. The results were subjected to a one-way analysis of variance (ANOVA) by the SPSS software and followed by a Tukey test to establish the difference between treatments.
Results
Molecular cloning, phylogenetic analysis, and multiple alignments of loach espl1 gene
The full-length cDNA of espl1 in loach is 6948 bp, and the open reading frame (ORF) of 6420 bp encoded 2139 amino acids with a molecular weight of 237.26 kDa, and the pI values was 7.00. This sequence comprises a 5′-UTR of 185 bp and a 3′-UTR of 343 bp, and the stop codon is TAG.
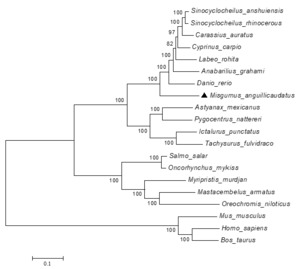
According to multiple alignments of the amino acid sequences, we found that the amino acid sequence of espl1 of loach is highly similar to the amino acid sequence of many other species concentrated in the C-terminal region (Figure 1) and contained three functional domain sequences: Peptidase_C50 (1744-2087aa), DTHCT (1407-1509aa), and TPR_12 (755-823aa), and also contained four conserved amino acid blocks (SVTRMPSL, LLFGCSSAAL, GNLWDVTDRD, GAAPIAYGLP). The box (E××R) was cleavage sites of espl1gene, and the arrow indicates the arginine residue after which cleavage occurs. The phylogenetic tree of the espl1 amino acid sequence was constructed based on the NJ method. The results showed that the mammals were gathered together as one, and all fish and invertebrates were brought together to form another branch (Figure 2).
Expression of espl1 gene in different tissues of adult loach
Analysis of espl1 gene expression level in adult loach by qPCR, espl1 expressed in all tested tissues of adult loach (Figure 3). In 14 tissues, Interestingly, it was detected that espl1 gene expression in adult loach was extremely abundant in the gonads compared to other tissues (P < 0.05) and rarely in the intestine and fin. The order was ovary > testis > > liver > muscle > kidney, brain > beard, gill, skin, heart, eye > fin, intestine.
Whole-mount in situ hybridization
Considering the high expression of espl1 in gonads of adult loach by qPCR, we want to know whether the function of espl1 is related to the development of the embryonic gonad during the early embryo development of loach. The expression of espl1 in the embryonic gonad was analyzed by whole-mount in situ hybridization. First, the expression level of espl1 mRNA in the early embryo development of loach was detected by qPCR (Figure 4). From 2 cell to early membrane release (2 h-24 h), the expression level of espl1 mRNA was very high compared to other stages of embryo, and the order was 7 h>2 h, 24 h>12 h>>36 h, 48 h, 60 h, 72 h, 84 h, 96 h. This result is consistent with the result of whole-mount in situ hybridization. From 2 h to 24 h, the espl1 expression was extremely comprehensive in the embryo of loach, and a special expression has been observed in the notochord since 36 h (Figure 4B-4C). At the same time, the expression of espl1 was so low from stages 36 h to 96h. However, no signal was detected in the embryonic gonads during the early embryo development of loach. Hence, It is likely that espl1 gene of loach is not expressed in the embryonic gonad; if expressed, it is weak and at an undetectable level.
espl1 specific expression in adult loach gonads
Although no signal of espl1 was detected in the embryonic gonads, the high expression of espl1 in the ovary and testis of adult loach was analyzed by qPCR and tissue in situ hybridization (Figure 5). Research has shown that vasa has been considered a specific molecular marker of germ cells, and the antibody of vasa is widely used to localize the primordial germ cells.14 As shown in Figure 5B and Figure 5D, the positive signals of espl1 and vasa genes were detected in the ovary and testis of the loach. The expression level of espl1 mRNA is highest in the oocytes stage Ⅳ and testis stage Ⅲ. As shown in Figure 5C, cleanly, different oocyte stage and testis stages of adult loach were observed by HE.
Effects of LHRH-A2 on espl1 expression in gonads of loach
To study the influence of LHRH-A2 on espl1 expression in loach during gonad development. Three groups of fish were injected with LHRH-A2; they were the control group (0μg/g), the low concentration group (0.1μg/g), and the high concentration group (0.2μg/g). qPCR was performed to measure espl1 mRNA expression after injection with different density LHRH-A2 (Figure 6). In the testes, espl1 significantly increased after 0.2μg/g and 0.05μg/g LHRH-A2 treatment at mRNA level (P < 0.05) (Figure 6A). Meanwhile, we detected the meiotic-related genes ldh and mapk. Compared with the control, both low and high-concentration LHRH-A groups significantly suppressed mRNA expression of ldh, and mRNA expression of mapk did not change significantly. Compared to the control, the mRNA expression level of espl1 was upregulated in ovaries, especially in the high-concentration LHRH-A2 group (injected with 0.2μg/g) (Figure 6B). While mapk showed a significant rise after LHRH-A2 injection, and mRNA expression of ldh increased significantly in the high concentrate LHRH-A2 group (Figure 6B). As shown in Figure 6C, there were different stage gonads with different densities of LHRH-A2. The results showed that injection of LHRH-A2 was beneficial to developing loach in gonads. in the control group, there were so many stage II ovaries and spermatogonium; in the high-concentration LHRH-A2 group, there were stage Ⅳ and Ⅴ ovaries and spermatoblast.
Discussion
In this study, we reported the first study of the espl1 function in loach. The full length of the espl1 gene of loach was first cloned and sequenced, and its open reading frame (ORF) is 6420 bp long and encodes 2139 amino acids. By multiple alignments and phylogenetic analysis of espl1, The large region of espl1 at the N-terminus does not show significant conservation between species, and its sequence similarity is limited to the C-terminal region. This is consistent with the findings in humans.15 Separase cleavage sites separate the conserved C-terminal catalytic domain of separase from the N-terminal portion,16 and all five cleavage sites that we identified in separase contain the consensus ExxR (Figure 1). Phylogenetic tree results showed that loach and zebrafish were one branch, while other fish were gathered into another branch. The results show that espl1 gene was closely related to zebrafish.
qPCR analysis revealed a strong tissue-specific expression of espl1 gene. This study showed the espl1 gene was expressed in 14 tissues of adult loach. Observably, the espl1 gene is highly expressed in the ovary and testis and is not expressed or has little expression in tissues such as the fin, intestine, beard, gill, skin, heart, and eye. So, we inferred that the high expression of espl1 in the ovary and testis suggests that espl1 plays an important role in developing loach gonads. At the same time, research shows that expression of the espl1 gene is associated with primordial germ cells (PGCs) in the early stages of mouse embryogenesis.10 To study the function of espl1 in the embryonic gonads during the early embryo development of loach, the expression of espl1 in the different stages of early embryo development of loach was analyzed by whole-mount in situ hybridization and qPCR. the espl1 gene is highly expressed at 2 h-24 h, and low expression at 36 h-96 h. A special signal was detected in the loach notochord by whole-mount in situ hybridization and rarely in embryonic gonads. These results indicate that espl1 plays an important role in the early embryo development of loach, especially regarding notochord formation.
Although the special signal also was not detected in the embryonic gonads, the expression of espl1 was detected in the ovary and testis of adult loach by qPCR. The espl1 gene was highly expressed in the oocytes stage Ⅳ and the testis stage Ⅲ. The Specific expression of espl1 in the gonads of loach shows that espl1 plays an important role in the ovary and testis of adult loach. separase encoded by espl1 gene is a key molecule for chromosome segregation and centrosome replication. In meiosis, the function of separase was to cleave cohesin at the onset of anaphase to release sister-chromatid cohesion.4 The function of the espl1 gene in the process of meiosis in loach is still unknown. Still, the high expression of espl1 in oocytes of ovarian development and testes indicates that espl1 is very important in meiosis.
To investigate the comprehensive expression of espl1 treated with two concentrations of LHRH-A2, it was used to stimulate loach. In the ovaries and testes, the injection of LHRH-A2 concentration increased the expression of espl1. These results suggested that LHRH-A2 induced meiotic in the gonad, and the high expression of espl1 shows that espl1 plays a role in the process of gonad meiosis in the loach.
In conclusion, we have cloned and characterized full-length cDNAs of espl1 for the first time in loach. Multiple alignments and the phylogenetic trees of espl1 were constructed here. Moreover, in loach, the espl1 gene was demonstrated to be tissue-specific and expressed in early embryonic development for the first time. More importantly, by whole-mount in situ hybridization, it was found that there was a significantly positive signal of espl1 in the notochord during the early embryo development of loach. Last but not least, our study showed that LHRH-A2 promoted the expression of espl1 in the testis and ovaries of loach. Therefore, we inferred that it had a distinct and important role in gonads and notochord formation during the early embryo development of loach. However, its specific function is still unclear and needs to be continued.
Acknowledgments
This work was supported by the Innovative Research Team of Dabie Mountains Fishery Resources Exploitation and Utilization in Xinyang Agriculture and Forestry University (XNKJTD-015); Analysis of the genetic structure of the population of green shrimp in Henan province based on whole genome resequencing technology (24B240001).
Authors’ Contribution – CrediT
Conceptualization: Hanjun Jiang (Lead). Methodology: Hanjun Jiang (Lead). Formal Analysis: Hanjun Jiang (Lead). Investigation: Hanjun Jiang (Lead). Writing – review & editing: Hanjun Jiang (Equal), Qianqian Huang (Equal), Xiaojuan Cao (Equal). Funding acquisition: Xusheng Guo (Equal), Jiahui Liu (Equal), Dexiang Feng (Equal). Resources: Xusheng Guo (Equal), Jiahui Liu (Equal), Dexiang Feng (Equal). Supervision: Xusheng Guo (Equal), Jiahui Liu (Equal), Dexiang Feng (Equal).
Competing of Interest – COPE
No competing interests were disclosed.
Ethical Conduct Approval – IACUC
In any case, we confirm that every effort has been made to alleviate the suffering of the test fish loaches, including the following details: All test fish were anesthetized before being sampled. We comply with the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article.



_and_.png)
._relat.png)




_and_.png)
._relat.png)
