Introduction
Global consumption of aquatic foods increased by 3.0 percent annually from 1961 to 2019, with the annual per capita consumption reaching a peak of 20.5 kg in 2019. To meet this demand, global aquaculture production has increased rapidly, from 21.8 million tons in the 90s to 214 million tons in 2020.1
At the national level, Vietnam is the world’s third largest exporter of aquatic products1; pangasius, tiger shrimp, and whiteleg shrimp are the main farming species.2 Exports of these products bring in large amounts of foreign currencies, making an important contribution to the country’s economic development, while aquaculture serves as the main livelihood for people in the Mekong Delta region and coastal localities.3,4 In recent years, Vietnam, like other major global seafood producers, has encountered numerous challenges in expanding aquaculture production. These challenges include environmental degradation such as mangrove depletion, declining water quality, and disease outbreaks in farming.4–6 The increasing scale of intensive farming leads to more frequent and difficult-to-control disease outbreaks.7 For pangasius, disease outbreaks in farms are mainly caused by environmental degradation, improper care, high stocking density, and low-quality fingerlings.8
Diseases have been reported to cause severe damage to aquaculture.9 Antibiotics and other veterinary drugs are commonly used in aquaculture to prevent and treat diseases.10 However, improper antibiotic usage can result in antibiotic-resistant bacteria or residues in processed products.11,12 Several alternative strategies, including vaccines, probiotics, herbal plants, and immunostimulants, have been proposed to prevent disease outbreaks and limit the use of veterinary drugs in aquaculture.13 As one of the best alternatives, probiotics have been strongly recommended because they contain secondary metabolites modulating biological functions, such as growth promotion, appetite stimulation, immune stimulation, disease resistance, antimicrobial activities, and extracellular enzymes that degrade organic compounds.14 Most of the probiotics proposed as biological control agents in aquaculture belong to the lactic-acid bacteria, the genus Bacillus, or the genera Pseudomonas and Burkholderia.14,15 Streptomyces have also been proven to be potential probiotics in aquaculture because they can produce antibiotics, antifungals, extracellular enzymes, siderophores, and anti-quorum sensing.16
This study was conducted to identify the indigenous Streptomyces strains with probiotic properties for potential use in aquaculture. These strains were tested on whiteleg shrimp to assess their ability to stimulate growth, improve feed efficiency, and provide protection against pathogens. The findings from this study will serve as a foundation for generating novel biological products to promote sustainable aquaculture practices.
Materials and Methods
Samples for isolating Streptomyces
The soil and sediment samples were taken at a depth of 10-15 cm from the surface and stored in 50 mL falcon tubes at 4°C for 72 h before being used to isolate Streptomyces. The sample collection sites are shown in Table 1.
Experimental shrimp
Whiteleg shrimp (L. vannamei) (61–70 days old, 16 g/each) were purchased from Nghe An, Vietnam. They were randomly tested for V. parahaemolyticus, and V. harveyi by polymerase chain reaction.
Bacterial strains
The bacterial strains used to evaluate the antagonistic activities of Streptomyces in this study were provided by the Institute of Biotechnology and Food Technology, Hanoi Open University. Of these strains, E. ictaluri E.HOU/AG21, A. hydophila A.HOU/DT14 were isolated from infected pangasius, and V. harveyi VH/BT12, V. parahaemolyticus V.HOU/BT01 were isolated from infected shrimp.
Isolating and screening Streptomyces
Soil or sediment samples were air-dried at room temperature for 7 days to remove moisture and reduce the number of bacteria (except for Streptomyces species). Then, they were crushed and sieved through a sterilization screen to remove large debris. After that, the samples were mixed with CaCO3 (10:1 w/w) in the sterile petri plates and incubated for 2 days at 28°C. Following, 20 g of sample-CaCO3 mixtures were suspended in 180 mL of sterile distilled water in the flasks on a rotary shaker at 200 rpm for 30 min. The pretreated samples were diluted up to 10−6. An aliquot of 0.1 mL of the diluted samples was then overlaid on starch casein agar (SCA-HiMedia) (contained 50 μg/ml of nystatin and 5 μg/ml of ampicillin) and incubated at 28°C for 7 days. The colonies of Streptomyces observed on SCA showed a tough or powdery texture and a dry or folded appearance, adherence to the agar surface, and branching filaments with or without aerial mycelia. These colonies were categorized into several colors, namely gray, white, red, yellow, green, brown, pink, orange, and violet (determined through comparison with the color charts of Inter-Society Color Council National Bureau of Standards- ISCCNBS)17; regarding morphological and biochemical characteristics, Streptomyces is gram-positive, filamentous, non-motile, and catalase-positive.18
Colonies of Streptomyces were purified using ISP2 agar (HiMedia). Purified Streptomyces were stored in ISP2 slants at 4°C or in 20% glycerol at -20°C.
Antimicrobial activity
The agar-diffusion method was used for antagonism detection. Streptomyces isolated were cultured on SCA for 7 days at 28°C. Four strains E. ictaluri E.HOU/AG21, A. hydophila A.HOU/DT14, V. harveyi VH/BT12, and V. parahaemolyticus V.HOU/BT01 were spread separately on tryptone soya agar (TSA-HiMedia). Subsequently, 6 mm plugs were excised from the Streptomyces plates, and placed on the TSA plates inoculated with those bacteria. The antimicrobial activity was monitored by measuring the diameter of the inhibition zone (halos; mm) around the agar plugs after 24 h incubation at 28°C.
Enzymatic activity
The agar-diffusion method was used to evaluate the ability of isolated strains to hydrolyze macromolecular polymers, such as proteins, starch, and cellulose.
Amylolytic activity: 6mm plugs were circled from the Streptomyces plates and placed on ISP2 agar (HiMedia) supplemented with 2% starch. The plates were incubated at 28°C for 3 days. After that, the surface of the agar was flooded with an iodine solution and then allowed to stand for a few minutes. The appearance of clear zones around the colony characterized amylolytic activity. The halo zone indicates the presence of amylase.
Proteolytic activity: Streptomyces isolated were screened for their proteolytic activity by using ISP2 agar containing skimmed milk 1%. The plates having the plugs of Streptomyces were incubated for 3 days at 28°C. After that, the plates were flooded with an HgCl2 solution (HgCl2 15 g, HCl 20 mL, and distilled H2O 100 mL). Protein hydrolysis appeared as a clear or white opaque zone around the colony.
Cellulolytic activity: ISP2 agar plates supplemented with 1% carboxy methyl cellulose (CMC) were used to measure the cellulolytic activity of Streptomyces. The plugs of Streptomyces were incubated on the above plates for 3 days at 28°C. After incubation, a lugol solution was dripped onto each of the plates. The halo zone around the colony indicates the presence of cellulase.
Salt and pH tolerance
The salt and pH tolerance of Streptomyces isolated was determined by growing in tryptone soya broth (TSB- HiMedia) at pH of 3, 4, 7.2, and 8.0 and salt concentrations of 0 ppt, 0.5 ppt, 2 ppt, 3 ppt and 5 ppt. The isolates were incubated at 28°C for 7 days. The presence or absence of growth was recorded after 7 days by UV-vis spectrophotometer at OD600.
Experimental design for evaluation of growth performance and feed conversion ratio
The experiments were performed in 100L glass tanks disinfected with 30 mg/L chlorine. Before the trials, L. vannamei was acclimated for 7 days under controlled temperature conditions (28 ± 0.5°C), salinity 15 ppt, and dissolved oxygen ≥ 4 mg/L; 30% of the water was changed daily.
The experiments lasted for 3 months (90 days), and experimental conditions were maintained as described above. The shrimp were fed with pellets (Cargill, HH724), which were supplemented with 108 CFU/g of isolated Streptomyces. In the control tank, the shrimp only consumed commercial pellets (without Streptomyces). The shrimp were fed 4 times daily to their needs. At the end of the experiments, the growth performance of the shrimp, such as weight gain (WG, %), daily weight gain (DWG, g/day), and feed conversion ratio (FCR) were calculated as follows:
WG (%)= 100×(final body weight (g)- initial body weight (g))/initial body weight (g);
DWG (g/day) = (final body weight (g)- initial body weight (g))/experimental days;
FCR= feed intake (g)/ total weight (g)
Protection activity of Streptomyces for the challenged shrimp
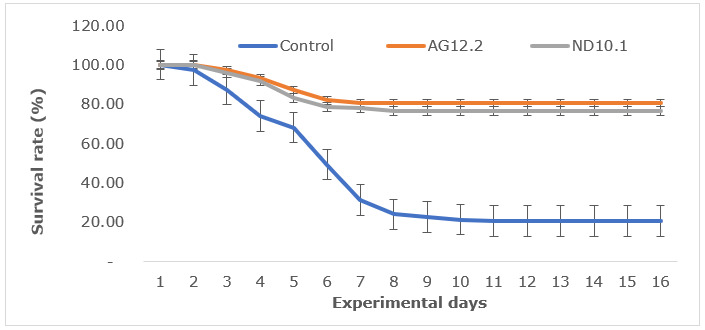
The experimental shrimp were fed with diets containing Streptomyces as described above. After 7 days of being fed with Streptomyces, the shrimp were challenged by being immersed in a suspension containing 106 CFU/mL of V. parahaemolyticus V.HOU/BT01. The shrimp in the Streptomyces control tank were fed daily on Streptomyces without challenge, while in the challenge control tank, the shrimp were challenged and fed with the diet without Streptomyces. All the tanks were continuously cultured for 15 days under the above conditions. The number of surviving shrimp was counted daily, with the survival rate being calculated using the following formula: survival rate (SR, %) = 100 x Wt/Wi, where Wi is the initial number of the experimental shrimp, Wt is the number of the shrimp that survived at the time of observation.
Sequencing for identification of Streptomyces
The genomic DNA of the isolated Streptomyces was extracted using a QIAamp DNA Blood Mini kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The eubacterial universal primers 27F: 5’-AGAGTTTGATCATGGCTCAG-3’ and 1492R: 5’-TACGGTTACCTTGTTACGACTT-3’ were used to amplify a product of approximately 1.5 kb of the 16S rRNA gene. The amplification reactions were performed in a final volume of 50 μL, which contained a DNA template, 2× PCR Master Mix solution (iNtRON, Korea), primers, and sterile distilled water. The thermal cycling conditions were as follows: initial denaturation at 94°C for 5 min; 35 cycles at 94°C for 60 s, 57°C for 60 s, and 72°C for 90 s; and a final extension at 72°C for 5 min. The amplified PCR products were checked on a 1% agarose gel, purified, and sequenced at the 1st BASE sequencing company, Seri Kembangan, Selangor, Malaysia. The sequences were subjected to BLAST (https://blast.ncbi.nlm.nih.gov/Blast) analysis for homology analysis. The relative sequences from the BLAST results were aligned and used to construct phylogenetic trees using CLC Workbench (version 22.0, Qiagen, Hilden, Germany).
Statistical analysis
All experiments were carried out in triplicate. Data were analyzed using SPSS 22.0 and one-way ANOVA. The result values were expressed as mean ± standard error, and differences were considered significant at the 0.05 probability level.
Results
Isolation and screening of Streptomyces
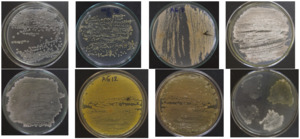
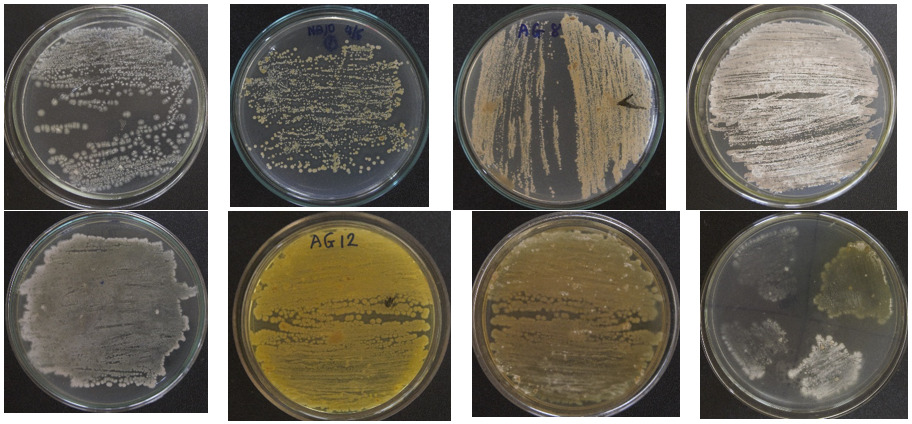
From 48 soil and sediment samples collected from different regions of Vietnam, a total of 58 strains were identified as Streptomyces because they have typical patterns, such as (1) smooth appearance colonies that later developed aerial mycelia which either appeared floccose, granular, powdery, velvet, or leathery; (2) varied colors of the substrate and aerial mycelia, including white, yellow-white, gray, brown, orange (Figure 1); (3) positive catalase and positive gram; and (4) filaments with branching mycelium. Eleven of those isolated Streptomyces strains were found to be resistant to four tested bacterial strains (pathogens in pangasius and shrimp), namely E. ictaluri E.HOU/AG21, A .HOU/DT14, V. harveyi VH/BT12, and V. parahaemolyticus V.HOU/BT01; among them, the five strains AG8.9, AG12.2, ND10.1, TB1.6, and ND5.3 were determined to be superiorly resistant to all four pathogenic bacterial strains with inhibition zones ranging from 18.70 mm to 28.93 mm (Table 2, Figure 2). Regarding extracellular enzyme activity, the 3 strains AG12.2, ND10.1, and TBL1 were found to have outstanding ability in the hydrolysis of starch, CMC, and skim milk; the size of hydrolysis zone was in the range of 19.60 mm to 32.13 mm (Table 3, Figure 3). Based on the evaluation of antibacterial activity and extracellular enzyme activity, Streptomyces AG12.2 and ND10.1 were initially identified as potential strains for application in aquaculture.
Salt tolerance and pH tolerance
Streptomyces AG12.2 and ND10.1 grew best at a salinity of 0 ppt to 2 ppt and pH of 7.2 to 8. Figure 4 shows that ND10.1 was more adaptive to salinity than AG12.2, and at the same time, it appears to be more acid-tolerant than AG12.2. Comparing the growth efficiency of these two strains at optimal salinity (0 ppt -2 ppt) and higher salinity shows that the biomass density of AG12.2 decreased by 2.43 times to 2.92 times corresponding to salinity of 3 ppt and 4.22 times to 5.09 times corresponding to salinity of 5 ppt; meanwhile, for ND10.1, biomass density decreased by 2.07 times to 2.34 times and 2.67 times to 3.02 times respectively salinity of 3 ppt and 5 ppt. Regarding pH tolerance, the biomass density of AG12.2 and ND10.1 decreased sharply under growth conditions at pH 3 and pH 4. Compare the growth efficiency of these two strains under the condition of optimal pH (7.2 - 8) and pH 3 and pH 4 shows that the biomass density of AG12.2 decreased by 2.82 times to 3.13 times at pH 3, decreased by 1.38 times to 1.53 times at pH 4; meanwhile, the biomass density of ND10.1 decreased by 2.42 times to 2.67 times and 1.40 times to 1.54 times at pH 3 and pH 4 respectively.
Growth performance and feed conversion ratio
Compared to the control group, the two strains AG12.2 and ND10.1 significantly improved growth performance in whiteleg shrimp, with the level of growth stimulation efficiency in shrimp of these two strains being equivalent (p > 0.05) (Table 4). Compared to the control group, weight gain and daily weight gain of shrimp fed with AG12.2 increased by 1.59 times and 1.64 times, respectively; meanwhile, in shrimp fed with ND10.1, these parameters increased by 1.61 times and 1.64 times, respectively.
The inclusion of Streptomyces in shrimp diets significantly reduced the feed conversion ratio compared to the control group (p < 0.05). Specifically, in groups of the shrimp fed with AG12.2 and ND10.1, FCR decreased by 1.3 times and 1.35 times, respectively (Table 4).
Survival rate of challenged shrimp
Both strains AG12.2 and ND10.1 showed the ability to protect whiteleg shrimp challenged with V. parahaemolyticus, and AG12.2 protected the shrimp more effectively than ND10.1. After 15 days of observation, compared to the control group, the survival rate of challenged shrimp fed with AG12.2 and ND10.1 increased by 3.9 times and 3.71 times, respectively (Figure 5).
Identification of the selected Streptomyces trains
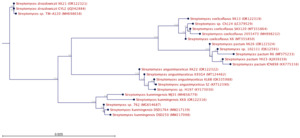
The amplified 16S rRNA gene sequences of Streptomyces AG12.2, ND10.1 were aligned with other sequences in Genbank using the BLAST tool (https://blast.ncbi.nlm.nih.gov/Blast). The nucleotide sequence data submitted with the Genbank accession numbers are shown in Table 5. A phylogenetic tree (Figure 6) was constructed according to the bootstrap test using the neighbor-joining algorithm with CLC Workbench 22.0 (version 22.0, Qiagen, Hilden, Germany). It shows the phylogenetic relationships of the two strains (XK8, the code for AG12.2 and XK 22, the code for ND10.1) with some other Streptomyces sp. strains in Genbank. Based on the nucleotide sequence data, colony characteristics, shape of spore chains, and biochemical characteristics of these two strains, the results show that XK8 (AG12.2) strain, collected at An Giang province, belongs to Streptomyces kunmingensis and XK22 (ND10.1) strain, collected at Nam Dinh province, belongs to Streptomyces angustmyceticus.
Discussion
Isolation and screening of Streptomyces
In this study, Streptomyces were isolated from all samples collected in several different ecological regions of Vietnam. The characteristics of salinity (0 ppt - 22 ppt), and pH (7.1 - 8.1) of the sample collection areas showed that Streptomyces are widely distributed in wetlands in Vietnam. The soil along riversides and sediments in shrimp ponds, fish ponds, and coastal estuaries are environments rich in organic matter, nitrogen, and total phosphorus, which are optimal for the growth of Streptomyces.19–21 The results of this study reveal that 18.97% of isolated strains were resistant to pathogenic bacteria in pangasius and shrimp, such as E. ictaluri, A. hydrophila, V. parahaemolyticus, and V. harveyi. Those strains can also hydrolyze starch, cellulose, and protein. Previous studies have also isolated and described the antibacterial, antifungal, and enzymatic activities of Streptomyces isolated from soil in different ecological regions in Vietnam20–22; however, this is the first time that Streptomyces have been isolated with a broad antibacterial spectrum against various pathogens that caused severe disease in shrimp and pangasius such as hemorrhagic disease and bacillary necrosis of pangasius, and luminous vibriosis, and acute hepato - pancreatic necrosis disease of shrimp; furthermore, they also produce a broad spectrum of extracellular enzymes that can hydrolyze several macromolecular polymers.
Salt tolerance and pH tolerance
As mentioned above, this study aims to find out Streptomyces strains with proper probiotic activity to develop biological products for application in aquaculture. Vietnam is a country that cultivates a diverse range of aquatic species that are farmed in inland, marine, and brackish waters23; therefore, the probiotic strains selected should be able to adapt to a multitude of environmental conditions, especially salinity and pH. In addition, developing probiotics for the feeding of livestock also needs to be considered in terms of pH tolerance in the digestive tract. Previous studies have reported that invertebrates lack gastric acid in the stomach, while the pH in the digestive tract of freshwater fish, which is normally in the range of 6.5 to 8.0, depends on the environmental temperature.24,25 The results of this study show that the two selected Streptomyces strains (AG12.2 and ND10.1) are adaptive to the salinity in the range of 0 ppt to 5 ppt and pH in the range of 3.0 to 8.0, in which they grow best at the salinity from 0 ppt to 2 ppt, and pH in the range of 7.2 to 8.0. Thus, these two strains are suitable for developing probiotics that are added to feed aquatic animals raised in Vietnam’s fresh, brackish, and coastal water.
Growth performance, feed conversion ratio, and survival rate of challenged shrimp
The present study on whiteleg shrimp shows that AG12.2 and ND10.1 could promote growth performance in shrimp, and at the same time, they also significantly improve feed efficiency. In the control group, WG, DWG, and FCR were 63.35%, 0.11 g/day, and 1.48, respectively, while the growth parameters of shrimp fed with AG12.2 and ND10.1 were as follows: WG was as high as 100.63% and 102.07%, DWG was as high as 0.18 g/day, FCR was as low as 1.14 and 1.10, respectively. These Streptomyces strains effectively protected whiteleg shrimp against V. parahaemolyticus; in the challenged control group (fed with diets without Streptomyces), the survival rate of shrimp was only 20.67%, while in groups fed with AG12.2 and ND10.1, the figures were 80.67% and 76.67%, respectively. Previous studies on the effects of Streptomyces on the growth in whiteleg shrimp have reported that DWG of shrimp fed with Streptomyces parvulus reached 0.12 g/day,26 and WG of shrimp fed with Streptomyces DH3.4 increased by 1.08 times.27 Reported by García-Bernal et al.28 showed that L. vannamei infected with V. parahaemolyticus was protected by Streptomyces RL8 and N7, the survival rate of shrimp fed with RL8 and N7 was 84.44 ± 5.7% and 57.77 ± 6.8%, respectively; meanwhile, the survival rate of challenged shrimp fed with the diet without Streptomyces was 11.11 ± 9.0%.
Identification of the selected Streptomyces strains
Through the application of 16s RNA gene sequencing, AG12.2 and ND10.1 were identified to belong to the species Streptomyces kunmingensis and Streptomyces angustmyceticus, respectively. These species have been shown to possess many beneficial characteristics, such as the ability to produce angustmycin with anti-mycobacterial and cytokinin activities, antifungal compounds, biosurfactant compounds, and antioxidant compounds.29,30 This finding shows that Streptomyces AG12.2 and ND10.1 are potential probiotics that cannot only be supplemented to the feeding regimen of aquatic animals to improve the growth performance, prevent diseases, and increase feed efficiency but also can be used to enhance the quality of aquaculture environment.
Conclusion
The typical characteristics of the two strains Streptomyces AG12.2 and ND10.1, which include (1) broad-spectrum antibacterial activity against many pathogenic bacteria, (2) ability to decompose macromolecular compounds, (3) salt and pH tolerance, (4) protective effect of infected whiteleg shrimp, and (5) improvement of growth performance as well as feed conversion ratio, show that these strains are excellent candidates for producing probiotics having various beneficial properties for application in aquaculture.
Acknowledgments
This research is part of the project DTDL.CN-31/21, funded by the Vietnam Ministry of Science and Technology. The authors would like to express our sincere thanks to Dr. Nguyen Van Quang and MA. Pham Hoang Anh for their provision of dedicated language support to complete the manuscript.
Authors’ Contribution
Conceptualization: Tam Pham Thi (Lead), Hien Nguyen Thi Thu (Equal), Phuong Vo Hong (Equal). Data curation: Tam Pham Thi (Lead), Hien Nguyen Thi Thu (Equal). Funding acquisition: Tam Pham Thi (Lead). Supervision: Tam Pham Thi (Lead), Phuoc Man Hong (Equal). Writing – original draft: Tam Pham Thi (Equal), Hien Nguyen Thi Thu (Equal), Hai Le Minh (Equal), Phuoc Man Hong (Equal). Writing – review & editing: Tam Pham Thi (Equal), Hien Nguyen Thi Thu (Equal), Phuong Vo Hong (Equal). Formal Analysis: Hien Nguyen Thi Thu (Equal), Binh Ta Thi (Equal), Thoa Vu Kim (Equal), Le Dinh Thi Thu (Equal). Methodology: Hien Nguyen Thi Thu (Equal), Huyen Vu Thi Bich (Equal), Mai Le Thi Tuyet (Equal), Hai Le Minh (Equal), Van Dao Thi Hong (Equal).
Competing of Interest – COPE
The authors of the research declare that there is no conflict of interest regarding financial support or personal relationships.
Ethical Conduct Approval – IACUC
The study was approved by the Animal Ethics Committee of Ha Noi Open University (protocol code 101/TB-HĐTVĐĐĐV, dated March 18, 2021).
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All data are available upon reasonable request.

_*e._ictaluri*_e.hou_ag21__(b)_*a._hyd.png)
_amylase__(b)_protease__(c)_cellulase.png)
_salt_tolerance__(b)_ph_tolerance.png)

_*e._ictaluri*_e.hou_ag21__(b)_*a._hyd.png)
_amylase__(b)_protease__(c)_cellulase.png)
_salt_tolerance__(b)_ph_tolerance.png)