Introduction
The Pacific salmon, also known as Oncorhynchus keta, is a member of Salmoniformes, Actinopterygii, and Salmonidae.1 It mostly consists of rainbow trout (Oncorhynchus mykiss), salmon (Oncorhynchus keta), and sakura masou formosanum (Oncorhynchus masou formosanum), which is a characteristic class of bottom cold water fish.2 The rainbow trout is native to the Kamchatka Peninsula and the Pacific coast of North America. Being commonly farmed cold water fish, they have a quick growth rate,3 delectable meat, a delicate taste, rich nutrition, and a high edible value. They are frequently employed as the top option for salmon sashimi fish and are well-liked by customers in the aquatic market.2 The salmonid farming scale is also expanding with the increase in salmonid demand. However, the spread of viral diseases, including Infectious hematopoietic necrosis virus (IHNV), Infectious pancreatic necrosis virus (IPNV), and viral hemorrhagic septicemia virus (VHSV), is a major threat to salmonid health.
IHNV is a linear single-stranded RNA virus that belongs to the family Rhabdoviridae and the genus Novirhabdovirus.4 Invertebrates, mammals, birds, reptiles, and fish are among the hosts of the three subfamilies of 40 species of slug viruses, according to the International Committee on Taxonomy of Viruses (ICTV) (https://ictv.global/report/chapter/rhabdoviridae/rhabdoviridae). The single member of the subfamily Gammarhabdovirinae is the Novirhabdovirus.5 Amend was the first to discover IHNV in sockeye salmon and rainbow trout from the Canadian region in October 1969. Through electron microscope observation, it was found to be similar to IPNV and VHSV morphology; it was temporarily given the moniker “hematopoietic organ necrosis virus” based on its clinical traits.6
IHNV is a viral disease that has spread widely in most countries around the world. The epidemiological characteristics of IHNV were mainly analyzed by its G protein. Five primary genotypes of IHNV, namely U-genotype, M-genotype, L-genotype, E-genotype, and J-genotype, can be distinguished using phylogenetic analysis of the IHNV-G protein genes.7,8 The IHNV infection was most prevalent in North America.9,10 U-genotype, L-genotype, and M-genotype were the predominant endemic strains in the northern United States.8,11 IHNV strains throughout Europe, including Iran, which is geographically close to Europe, predominate in the E-genotype.9 J-genotype predominates in China,12 Japan, Korea,13,14 and other Asian countries.12 In Japan, the M-genotype was also reported, and the observed time point was earlier than that of the J-genotype.
China has experimented with breeding rainbow trout and other salmon since the 1990s. The Heilongjiang Fisheries Research Institute in China first detected the virus in the high-density breeding technology test tank of juvenile rainbow trout in the Bohai cold water fish Test Station in 1985.15,16 IHNV has since expanded throughout China (Figure 1). Since then, IHNV has been reported in more than ten provinces. This study analyzed the pathogenic characteristics of the IHNV-SCCD strain. Through the proximity methods, 100 IHNV strains, including IHNV-SCCD, separated from China, were constructed the Phylogenetic trees, and analyzed IHNV’s epidemiology in China. This provides some references for future IHNV identification and epidemiological analysis in China.
Materials and Methods
Experimental animals and cells
The diseased rainbow trout were collected a rainbow trout farm from Chengdu, Sichuan Province, China. The body length was 6 to 8 cm. The healthy rainbow trout were subjected to a week-long acclimation period in aerated freshwater tanks maintained at 15 °C. The Epithelioma Papulosum Cyprinid (EPC) cells were cultivated in a medium 199 (Gibco) solution containing 10% fetal bovine serum and maintained at a temperature of 25°C.
Sample collection and pathological examination
To ensure the life of diseased rainbow trout before sampling, fish were kept in a tank with an oxygen pump and transported to the lab. Clinical symptoms were noted and photographed. Tissue samples from the liver, spleen, kidney, and gills were collected and kept at -80 °C. The tissue was collected and placed in formalin. The pathological tissue was observed after washing, dehydration, penetration, embedding, and batik removal.
PCR detection
According to its instructions, total RNA was extracted from the diseased fish using the RNA Rapid extraction kit (Tiangen, China). The primer sequences of IHNV17 and IPNV18 were as follows. (IHNV-Forward: GTTCAACTTCAACGCCAACAGG, IHNV-Reverse: TGAAGTACCCCACCCCGAGCATCC). (IPNV-Forward: CCGCAACTTACTTGAGATCCATTATGC; IPNV-Reverse: CGTCTGGTTCAGATTCCACCTGTAGTG). The size of the amplified characteristic products was 371bp and 206bp, respectively. Both of the annealing temperatures were 60℃. Purified PCR products were sequenced at TsingKe, China, and compared using the blast function of the NCBI (Basic Local Alignment Search Tool) system.
Virus isolation
The liver, spleen, and kidney tissue samples were mixed and ground into tissue fluids using a tissue grinder. 2% fetal bovine serum-containing medium was re-suspended in a 1:100 ratio (weight to volume ratio). The antibiotic to antifungal ratio was 100:1 (volume ratio). The tissue suspension was freeze-thawed three times in a -80 ℃ refrigerator. Freshly made monolayer epithelial tumors were infused with the tissue suspension after it had been filtered via a 0.22 μm sterile filter. In a medium containing 2% fetal bovine serum, the supernatant was cultured at 18°C. After blind passing every week, the cytopathic effect (CPE) was observed every day. The cells with CPE were digested with trypsin and fixed in PBS containing 2.5% glutaraldehyde for electron microscopy (Lilai, China).
Infection experiment
Healthy juvenile rainbow trout were randomly divided into 6 groups of 20 fish per group and kept at 15 ° C water temperature for adaptation for one week. The fish was challenged by intraperitoneal injection at doses of 6.48×104TCID50, 6.48×103TCID50, 6.48×102TCID50, 6.48×101TCID50 and 6.48×100TCID50, respectively. The control group was injected with the same volume of PBS. The TCID50 of IHNV-SCCD was calculated according to Koch’s method.
Complete genome sequencing
Total RNA was extracted from the cells with typical CPE using the RNA Rapid extraction kit (Tiangen, China) according to its instructions, then reversed transcription into cDNA using the Vazyme reverse transcription kit (Tiangen, China) and sent to Shenzhen Tappu Gene Technologies Co., LTD. for metagenomics analysis. After cDNA fragments were fragmented by a Covaris M220 instrument, both ends of the fragmented cDNA were subjected to repair reactions and connected to sequencing adapters using the TruSeq™ DNA Sample Prep Kit, and libraries with sizes outside the 200-500bp range were removed by magnetic bead screening after index addition. The cDNA was amplified to a single strand by bridge PCR experiments using TruSeq PE Cluster Kit. Using the Illumina Novaseq6000 PE150 in-depth sequencing. Using paired-end (PE) and bbmap raw data for quality inspection and after pruning, Lenovo was used for Lenovo assembly, and SPAdes and SOAPdenovo software were used to assemble the next-generation contigs. The contigs obtained from the above assembly were compared with the virus-NT database by BLAST(V2.10.0+).
Genome analysis
Comparing, downloading, and sequencing the nucleic acid sequence took place at NCBI. Download in FASTA format. Sorted sequences were sorted using clusters (codons). The option of MEGA 11.0 program to set bootstrap copy to 1000.19 The homology of the nucleotide and amino acid sequences was compared using MEGA 11.0’s Align option. Using IQ-tree (V2.1.2) software of Maximum Likelihood method (Maximum Likelihood, ML), set the bootstrap to 1000 to build an evolutionary tree nodes support value obtained. The tree was visualized using the online software iTOL (https://itol.embl.de/).
Results
Clinical and anatomical symptoms
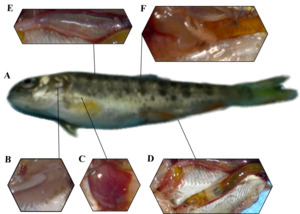
The rainbow trout infected with IHNV looks black (Figure 2-A). White gill filament and anemia (Figure 2-B), bleeding on the liver surface (Figure 2-C), thinning and transparent intestinal wall after dissection (Figure 2-D). The swim bladder turned red and exhibited bleeding (Figure 2-E) and spot bleeding of adipose tissue (Figure 2-F).
Histopathology observation
Histologically, the splenic tissue structure was unclear at low magnification; serous exudates were visible and the number of lymphocytes was decreased (Figure 3-A1). The splenic parenchyma cells were necrosis at high magnification, with nuclear fragmentation and lysis (Orange range) (Figure 3-A2). The renal interstitial tissue exhibited edema and reduced cellular components, with almost necrotic and lytic renal interstitial cells(Rad range) and many inflammatory cell infiltration (Figure 3-B1). The glomerular structure was disordered (Black arrow) (Figure 3-B2). The liver cells degenerated and the structure was disordered (Blue range) (Figure 3-C). The gill lamellae were hyperplastic and confluent at the end (Black range). The epithelial cells of the gills increased (Yellow range) (Figure 3-D).
Virus isolation
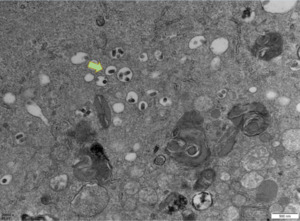
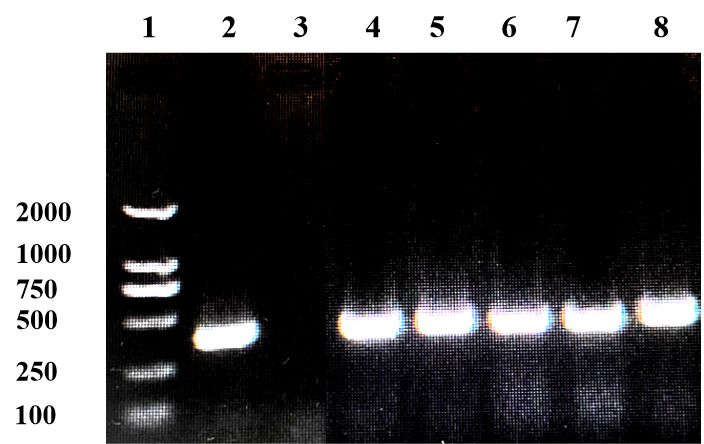
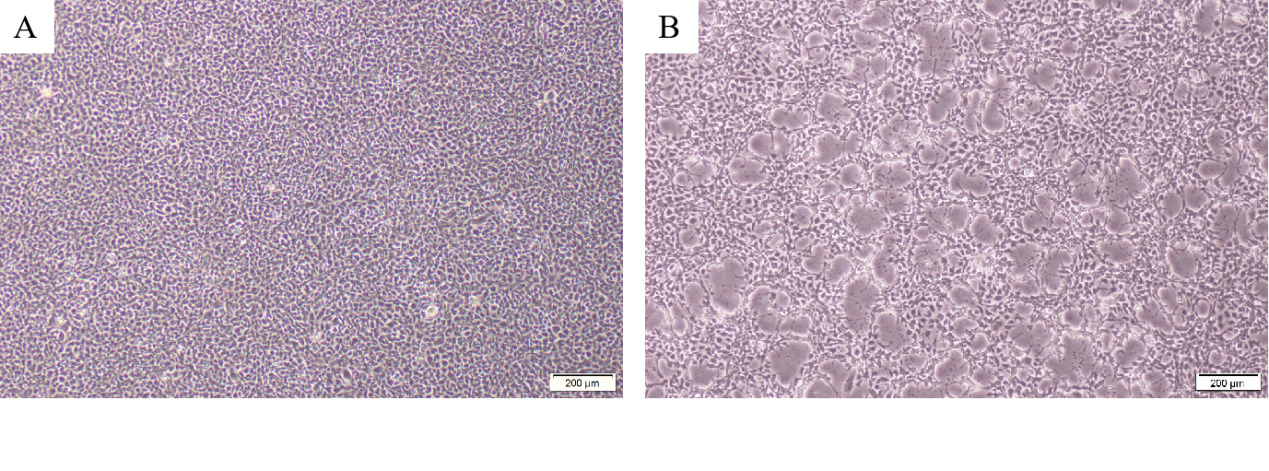
PCR results revealed that five samples in this study were infected with IHNV, which had amplified the characteristic products with a size of 371bp (Figure 4). The result of IPNV was negative (data not shown). After the tissue fluid was inoculated with fresh cells, a typical CPE phenomenon was observed on the fifth day. Microscopically, the control EPC remained normal, with tight intercellular connections and no empty spots (Figure 5-A). After IHNV infection, many plaques were formed at low magnification (Figure 5-B), and cells near the plaque tended to become round and shrink. The cells with typical CPE characteristics were sent for transmission electron microscopy, which revealed a large accumulation of virions in the cytoplasm. It can be seen that the IHNV virus particles present a typical bullet head shape with a length of approximately 130nm and a width of about 80nm (Figure 6).
Infection experiment
Rainbow trout were infected by intraperitoneal injection with different concentrations of IHNV-SCCD (Figure 7). In the highest dose group, death began to occur 24 hours post-infection, and the cumulative mortality reached 100% at 5 days. In the 3.24×104TCID50/ mL dose group, the death began to occur on the 2nd-day post-infection, the mortality peaked at 3-5 days post-infection, and the cumulative mortality reached 100% at 8 days. In the intermediate-dose group, death occurred at 3 days post-infection, and the cumulative mortality reached 65% at 2 weeks. In the 3.24×102TCID50/mL dose group, death began to occur about 4-5 days post-infection, and the cumulative mortality rate was 30% at 2 weeks. The cumulative mortality in the lowest dose group was 10%. No clinical signs or deaths occurred in the PBS group. The TCID50 of IHNV-SCCD was 8.61×102 TCID50/ mL.
Complete genome sequencing and analysis
The IHNV strain isolated in this study was named IHNV-SCCD, and the whole genome sequence has been uploaded to NCBI (Genbank No. OQ801357). The total length of the nucleic acid sequence of the IHNV-SCCD strain was 11161nts. Except for the 3 ‘leader region and the 5’ Trailer region, the middle sequence encodes six proteins: Nucleoprotein (N), Phosphoprotein (P), Matrix protein (M), Glycoprotein (G), Non-Virion protein (NV) and Polymerase(L)(Figure 8). The proteins werre separated by UTR region. The CDS area of Nucleoprotein is 1176 bp, with 391 amino acids; Phosphoprotein has a CDS area of 693 bp, with 230 amino acids; Matrix protein has a CDS area of 588 bp, with 195 amino acids; Glycoprotein has a CDS area of 1527 bp, with 508 amino acids; Non-Virion protein has a CDS area of 336 bp, with 111 amino acids; and Polymerase has a CDS area of 5799 bp, with 1932 amino acids, as summarized in Table (Figure 8).
The complete genome of 13 IHNVs isolated from different countries and 4 strains belonging to other members of the Rhabdoviridae family were analyzed . IHNV-SCCD belongs to the Rhabdoviridae family and is clustered together with IHNVs from all over the world (Figure 9). This observation effectively rules out the possibility that IHNV-SCCD is a member of other Rhabdoviridae viruses such as SVCV (DQ491000), VHSV (KY359354), SHRV (AF147498), and HRV (AF104985) (Black range). The IHNV strain from Shaanxi Province (Genbank No.MH628874) had the highest similarity value of 79 with IHNV-SCCD. In addition, the IHNV strain with high similarity were from China (Genbank: MN225937; MT242597; JX649101; MF509529; MH374162; KJ421216) (Green range). The strain was quite different from the IHNV strain from European countries such as the United States and Italy (Blue range). Notably, the sequences of IHNV strains from the United States (L40883 and HM461966) and European countries such as France (MN856664) and Italy (MK829714 and MN914162) also exhibited regional divergence.
Protein gene analysis
Except for glycoproteins, the remaining five proteins of IHNV-SCCD strain were compared and analyzed (Figure 10). The nucleoproteins displayed distinct regional characteristics. Similar strains from neighboring countries were clustered together, with IHNV-SCCD exhibiting the highest degree of similarity (94%) to the SX1704 strain (MH629974) isolated from Shaanxi Province, China, followed by other Chinese strains. Interestingly, the Phosphoprotein of IHNV-SCCD was classified at a higher level than many Chinese IHNV isolates, indicating an earlier appearance. The Polymerase of IHNV-SCCD shared complete nucleotide sequence identity with the SX1704 strain from Shaanxi Province, China and had a similarity of 100%. The BjLL strain, on the other hand, was more closely related to IHNV isolates from France, Italy, and other European countries, as well as the United States. The Matrix protein of IHNV-SCCD strain was located at the end of the phylogenetic tree and belonged to SX1704 strain, which in turn had the same nucleotide sequence with HLJ-09strain (JX649101) and 20101008 strain (KJ421216). The Non-Virion protein (NV) of the IHNV-SCCD strain belonged to the Chinese branch. Except MsYy1063b strain (MF325027), the Non-Virion proteins of all IHNV strain from Korea and China belong to the same branch. Notably, both the Matrix protein and Non-Virion protein of the BjLL strain were more similar to those of the US strain.
Epidemiological analysis of IHNV in China based on glycoprotein
IHNV can be classified into five types based on their glycoprotein sequences: U, E, M, L, and J. E genogroup was the main epidemic in Europe, J genogroup was the main epidemic in Asian regions such as Korea and Japan, among which U genogroup isolates also appeared in Japan, M, and L genogroup were mainly circulated in the United States, and U genogroup isolates also existed in the United States. As the primary reference for IHNV evolutionary typing, the glycoproteins of 33 IHNV strains from different countries were aligned and analyzed (Figure 11). Among them, there were three strains of L genogroup, which were from the United States; Five strains of E genogroup from Europe; Four strains of M genogroup, from the United States; There were nine U genogroup strains, four from the United States and five from Japan. In addition, there were 22 strains of J genogroup, all of which were from Asia, and 7 strains of J-S subgroup, including 5 strains from Japan and 2 strains from South Korea. There were 15 strains of J-N subgroup, including 5 strains from China, 5 strains from Japan and 5 strains from South Korea. It was found that the glycoprotein belongs to the typical J genogroup, which is closer to the Nagano subtype of the J type. It was closest to the Chinese strains. J-N subgroup were grouped into separate clusters according to national geographic location.
In order to understand the genetic and evolutionary relationship of IHNV in China, the glycoprotein sequences of 99 strains of IHNV from China were analyzed and compared (Figure-12A). By province, 40 strains from Liaoning Province, 13 from Heilongjiang Province, 5 from Jilin Province, 4 from Shandong Province, 5 from Beijing Province, 12 from Gansu Province, 5 from Hebei Province, 6 from Sichuan Province, 6 from Xinjiang Province, 3 from Qinghai Province, 1 from Shaanxi Province, and 5 from Yunnan Province were included. The IHNV isolates were analyzed according to the three regions of the northeast (Figure-12B), southwest (Figure-12C) and northwest (Figure-12D). The genetic distances of IHNV isolates within each province (Intra-province) and the genetic distances between the isolates in each province and IHNV-SCCD were calculated (Figure 13). After nearly four decades of evolution in China, IHNV has formed many different clades. In this system, IHNV-SCCD was clustered with YZH140519 (KM411979) and QQJ16(MN475923) from Sichuan Province. The IHNV-SCCD strain had the closest genetic distance with the IHNV strains isolated in Sichuan province, and the nucleic acid diversity was 2.0%-3.7%. The IHNV-SCCD strain had the longest distance with the IHNV strains isolated in Xinjiang province, and the nucleic acid diversity was 4.0%-4.8%. BjLL (KP216199) isolated from Beijing and LN12-17(MF802755) isolated from Liaoning Province were identified as MA and MN subtypes of U-genotype, respectively. The number of strains in Liaoning province was the highest, and the genetic relationship was close to that in other provinces. In the green part, the branches are long in length and distantly related. The strains in Heilongjiang province evolved rapidly and were closely related. The strains from Jilin province were more closely related to those from Liaoning province than those from Heilongjiang province. The Sichuan strains were closely related to the Yunnan strains, and some Sichuan strains were located in the upper level of Yunnan strains, and some strains were distantly related. The genetic relationship between Yunnan strains and Xinjiang strains was high, up to 75. BJ1401 and BJ1402 were closely related to SC15 and LQN131107. QH17 from northwest China was clustered with the SX1704 strain from Shaanxi province, while QH15 and QH16 were clustered with some strains from Gansu province, and the remaining strains from Gansu were clustered with the YN15 strain from Yunnan province, with a genetic relationship of up to 86.
Discussion
Over the few years since the 1960s, when salmon were introduced to China, IHNV has spread rapidly throughout the country .20 The main breeding areas of rainbow trout in China, the northeast and southwest regions, have been particularly affected by IHNV.21 In addition, IHNV has also been detected in the northwest and North China. Subtype J has been identified as the predominant type of IHNV in China, showing an evident evolutionary relationship. In the early years, China introduced large quantities of rainbow trout fry from Japan and the United States, and some researchers speculate that IHNV in China might have originated from Japan. However, its origin remains unclear due to the age of the first IHNV isolated in China and the lack of molecular analysis. In this study, we isolated an IHNV strain in Sichuan Province, China, the sixth strain isolated in Sichuan Province recorded in NCBI. The remaining fifth strain were LQN131107(KJ441079) in 2013,SC15(MH170337) in 2014, SC14-1811 (MF802781) in 2014, YZH140529 (KM411979) in 2014 and QQJ16 (MN475923) in 2016. Notably, as depicted in Figure 10, the aforementioned three IHNV strains were found to be closely related to the strain isolated in this study, with YZH140529 being the closest, thus providing strong evidence for their evolutionary relationship.
The kidney and cephalic kidney were the primary target organs of IHNV in infected fish.22 Clinical studies have shown that the clinical symptoms of IHNV include a black body color, necrosis of the gill and liver,23 spleen, kidney, and other organs, loss of hematopoietic function, overall anemia, and edema in infected fish. Histopathology of infected fish typically reveals necrosis and edema of histopathocytes, resulting in the enlargement of the intercellular space.23,24 The intestinal tract was also commonly affected,22 with the intestinal wall becoming thin and transparent and the absence of the intestinal muscle layer observed in histopathology. In the present study, the gill filaments of infected rainbow trout were white, indicating anemia. Histopathology revealed a decreased number of blood cells, edema-induced loss of the respiratory epithelium of the branchial lamella, and a larger-than-normal basic structure of the branchial tissue, which causes dyspnea and hypoxia in fish and may contribute to their death. Hemorrhagic spots were observed on the surface of the liver, which was in the early stage of IHNV-induced liver lesions. Although bleeding was present, there was no extensive loss of hematopoietic function. In histopathology, a few nuclei of hepatocytes were broken and dissolved, but some intact cells still existed. Changes in the gut are also typical. Histopathology revealed the complete disappearance of the intestinal muscle layer, obvious edema, and shedding of the intestinal mucosa. Additionally, tissue cells of the kidney and spleen were necrotic, with broken nuclei and incomplete tissue forms. Thus, it is inferred that IHNV primarily targets organs with strong hematopoietic function, such as the kidney and spleen, with liver lesions occurring later. The gill tissue lesion and the disappearance of hematopoietic function make breathing difficult for sick fish and also negatively impact their oxygen uptake capacity.
In the same family as SVCV and VHSV, IHNV possesses a bullet-like or rod-like virion structure.25 IHNV is capable of replicating26 in EPC cells, rainbow trout gonadal cells (RTG-2),27 Chinensis chinensis gonadal cells (CHSE-214), and bluegill sunfish cell lines (BF-2) in vitro at an optimal temperature range of 15-18℃.28 In the current study, IHNV was isolated and cultured in EPC and RTG-2, which produced typical cytopathic effects (CPE) and yielded positive cell supernatant test results by PCR. Samples from both tissue and in vitro cell cultures were fixed with 3% glutaraldehyde and observed under electron microscopy. However, no virions were observed in the tissue samples, while large accumulations of cell particles were observed in the cell samples, mainly in the cytoplasm. It was speculated that the amount of virus present in the tissue samples was insufficient to be visible under the electron microscope.
Up to now, 36 pieces of data on the whole genome of IHNV have been recorded in NCBI, and 7 pieces of data have been from China.12 To enhance our understanding of Chinese IHNV isolates, this study utilized the Illumina Hiseq platform to construct RNA genome paired-end libraries and sequence the entire genome. The resulting data was then uploaded to the NCBI official website. Our analysis revealed that IHNV-SCCD has a total gene length of 11,161 nucleotides and six coding regions, which encode six proteins.
Phylogenetic analysis compared IHNV-SCCD with 12 IHNV strains from four countries, including China. The strain most closely related to IHNV-SCCD was SX1704 (Genbank: MH629974), which was isolated from Shaanxi Province, China. By analyzing the six proteins, our study found that the nucleoprotein and polymerase of IHNV-SCCD were most similar to those of SX1704. Interestingly, the matrix protein, phosphoprotein, and non-virion protein of SX1704 clustered with IHNV-SCCD, suggesting a close genetic relationship between the two strains. IHNV-SCCD was isolated in 2017, and SX1704 was isolated in 2018, which were the first confirmed cases in Shaanxi Province. In China, Shaanxi Province and Sichuan Province are neighboring provinces that are bordered by land. All evolutionary trees were analyzed in combination; our findings suggest that IHNV transmission in Shaanxi Province may have originated from Sichuan Province.
Glycoprotein was the basis of IHNV virus classification. At present, J-type IHNV is mainly circulating in China, and two U-type IHNV strains, respectively BjLL strain (MF509592) and LN12-17 strain (MF802755), were isolated in 2012.29 To better understand the genetic relationships among Chinese IHNV strains,98 glycoprotein sequences of IHNV strains from China available in NCBI were compared and analyzed. The isolates from different provinces were analyzed by region.
First of all, the three eastern provinces were the first provinces in China to try salmon trout breeding. Heilongjiang province was the province that reported IHNV for the first time in China. Liaoning Province was the province that recorded the most IHNV strains in NCBI, but the number of reports in Jilin Province was smaller. The sample size in Liaoning Province was large, and the evolutionary direction had two major branches related to most of the strains in China(Figure 11-B). The green part had many long branches, indicating that IHNV might have experienced virus resistance in the salmon trout of Liaoning Province for a period of time and then began to spread widely in Heilongjiang and Liaoning Province. The strains in Heilongjiang province were closely related and spread quickly. Corresponding to this conclusion, the genetic distance of the strains in Liaoning province was the farthest, twice that of Heilongjiang province, and the lowest in Jilin province.
The IHNVs in Xinjiang have the closest genetic relationship with those in Yunnan (Figure 11-A); therefore, the Xinjiang region was added to constructing the evolutionary tree in the Southwest region(Figure 11-C). Among them, the mainstream of the Yangtze River runs through Sichuan and Qinghai provinces, which share water resources, and the salmon trout in Yunnan province are mainly distributed in the northeast and northwest of the province, where cold flowing water is rich. According to the Figure, LQN131107, SC14, and SC14-1811 strains from Sichuan province were found to be at the upper level of Yunnan and Xinjiang strains. Combined with their geographical location, it is speculated that some of the Yunnan strains may have been imported from Sichuan. Yunnan was located in the upper level of Xinjiang, and it was speculated that some of the strains in Xinjiang were introduced from Yunnan, which may be caused by the fact that Xinjiang belongs to a dry and cold area, which is not suitable for large-scale production, there are many self-employed people, and the way of purchasing fish fry is not standardized. To understand the source of Sichuan strains, we combined and analyzed some sequences from Sichuan and Beijing and Liaoning and Heilongjiang(Figure 11-D). Among them, SC15 is the original strain of the last part of the evolutionary tree, and in this Figure, it is located at the next level of the partial Beijing strain, but SC14-1811 is located at the next level of the partial Beijing strain. BJ13 was clustered with IHNV-SCCD, QQJ16, and YZH140529, so we speculated that Sichuan and Beijing strains were closely related, each promoting the evolution of strains in the other province.
Finally, we analyzed the strains from Qinghai, Gansu, and Shaanxi provinces in the northwest region and LN12-14 and YN15, which are most closely related to GS14-MXP (Figure 11-E). Among them, SX1704 is the only one recorded in Shaanxi province. SX1704 was most closely related to QH17, but the two clades independently clustered into one cluster, separate from QH15 and QH16. The Gansu strains were also mainly divided into two clusters. QH15 and QH16 were more closely related to one cluster of GS strains, while the other part of Gansu strains was more closely related to YN15. This suggests that there are two major evolutionary clusters in Gansu, one related to Qinghai and one related to Yunnan. There were also two clades in Qinghai, and the genetic distances of these two clades were longer than those of Gansu. This is also consistent with the conclusion of the genetic distance(Figure 12).
Based on the genetic distance, Liaoning and Heilongjiang provinces of China had the largest nucleic acid diversity of 8.8% and 4.9%, respectively. Yunnan and Xinjiang had the slowest evolutionary rates of 0.1% and 0.6%, respectively. In addition to this, it is worth noting that SD13a, SD14a, and SN1203 strains from Shandong, which are more distant from the other Chinese strains, still belong to the Nagano subgroup of type J. This may be related to the fact that Shandong province faces Japan across the sea, and IHNV can also infect salmonid Marine fish.
In this study, one IHNV strain was isolated, and its whole genome was sequenced. It was then compared with the currently reported IHNV strains in China, and the prevalence of IHNV in China was roughly analyzed by province. However, due to the limitation of calculation methods, the conclusions obtained in this study are not systematic enough. This is also the main direction in which to improve in the future.
Ethics statement - IACUC
The Animal Care and Use Committee of Sichuan Agricultural University reviewed and approved the animal study.
Acknowledgments
This work was supported by the Sichuan Science and Technology Department (grant number 2021YFN0123) and the Chengdu Science and Technology Project (2022-YF05-00641-SN).
Authors’ Contribution
Data curation: Yankai Li (Equal), Wenqian Li (Equal), Jiaxing Liu (Equal), Shuhan Li (Equal), Qiunan Li (Equal), Defang Chen (Equal), Wenyan Wei (Equal). Writing – original draft: Yankai Li (Lead), Wenqian Li (Equal). Writing – review & editing: Yankai Li (Equal), Wenyan Wei (Equal). Conceptualization: Qian Yang (Equal), Yongheng Zhou (Equal). Investigation: Yang He (Equal), Yi Geng (Equal). Validation: Yang He (Lead). Formal Analysis: Yang Ma (Equal), Shuhan Li (Equal), Yongheng Zhou (Equal). Methodology: Yang Ma (Lead). Resources: Xiaoli Huang (Equal), Yi Geng (Equal), Wenyan Wei (Equal). Software: Xiaoli Huang (Lead). Funding acquisition: Ping Ouyang (Lead).
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All are available upon reasonable request.






__.png)

_(figure_10a)__phosphopr.jpeg)
_sequences_._according.jpeg)
_sequences_with_ihnv_stra.png)






__.png)

_(figure_10a)__phosphopr.jpeg)
_sequences_._according.jpeg)
_sequences_with_ihnv_stra.png)
