Introduction
Vietnam’s favorable conditions support significant aquaculture development, particularly shrimp farming. White leg shrimp (Litopenaeus vannamei) and black tiger shrimp (Penaeus monodon) are major aquatic export products. However, Desulfovibrio spp., H2S-producing bacteria, poses a significant threat to this industry. These sulfate-reducing bacteria thrive in anaerobic environments, converting sulfate to hydrogen sulfide.1 The resulting H2S, a metabolic byproduct, is harmful at high concentrations. It is carcinogenic, toxic to intestinal cells, inhibits oxygen binding to cytochrome c, disrupts oxidative phosphorylation and ATP formation, and can mutate DNA and disrupt protein structures. The increasing incidence of bowel diseases in Western countries highlights the importance of research on managing Desulfovibrio spp. and its impact on gut health.2 In shrimp ponds, Desulfovibrio spp. proliferate in high-organic-matter environments, such as those with bottom residues, sludge, and untreated crop liners. This leads to toxic gas accumulation, particularly H2S, under the pond bottoms.2 Waste and leftover feed contribute to anaerobic conditions, resulting in foul odors and potential shrimp mortality. Effective pond management is crucial to prevent organic pollution and maintain shrimp quality, as neglect leads to substantial farmer losses.3
Antibiotic use in aquaculture results in antibiotic residues in water and sludge, promoting antibiotic-resistant bacteria. Bacteriophages, viruses that infect bacteria, offer a potential alternative for controlling bacterial diseases. Discovered by William Twort in 1915 and Felix d’Herelle in 1917,4 bacteriophages are a promising biocontrol method in aquaculture, minimizing environmental and human impacts. This approach, initially used in Japan,5 has gained significant scientific interest. Previous studies have demonstrated the lytic activity of bacteriophages against various Desulfovibrio species.6–10 This study aimed to identify and evaluate potential bacteriophage strains for inhibiting Desulfovibrio spp. in shrimp ponds.
Materials and Methods
Materials
Desulfovibrio sp. bacteria were obtained from the Can Tho University project: “Application of bacteriophages to treat toxic gas-producing bacteria causing decreased oxygen levels in shrimp ponds” (Project code: TSV2021-156). The genus-level identification, based on morphological, physiological, and biochemical characteristics, had been reported by Van & Thu.11 This study further identifies the species level using 16S rRNA gene sequencing. Bacteriophage strains ST231, ST22, φMix 1 (M1: ST231 + ST22), φS8, φS9, and φMix 2 (M2: S8 + S9) (isolated in Tra Vinh province) which were provided by the Molecular Biology Laboratory, Institute of Biotechnology and Food Technology, Can Tho University.
Isolation and Identification of Desulfovibrio spp. bacteria by 16S rRNA gene sequencing
Bacterial DNA was extracted, and PCR was performed using 16S rRNA primers. PCR products were electrophoresed on agarose gel to confirm the expected ~1500 bp band. Samples with clear bands were sent to the 1st BASE laboratory for sequencing. The 16S rRNA gene sequences were compared using the BLAST program in the NCBI database to identify the bacterial strains.
Investigation of the inhibitory effects of bacteriophage on Desulfovibrio spp. bacteria in the laboratory and 2-liter jar models
Seven liters of shrimp pond water were divided into seven 1-liter plastic jars. Water parameters (dissolved oxygen (DO), H2S, and NH3/NH4+ levels) were monitored. Pre-inoculation agar plating was performed. Each jar received 2 mL of the Desulfovibrio vulgaris strain 12Đ (species identified through 16S rRNA sequencing), 7 mL of TSB medium, and 50 mL bottom sludge. Then, 250 μL of each bacteriophage (or 125 μL of each phage for φMix) was inoculated. Post-inoculation agar plating was performed at 3, 6, and 18 hours at a 10-4 dilution. Water parameters were monitored at 3 and 18 hours. Dissolved oxygen (DO), H2S, and NH3/NH4+ levels were measured using Sera test kits.
Evaluation of the effects of bacteriophages on colony morphology of Desulfovibrio spp.
Using a plaque assay method, 200 μL of control (shrimp pond water inoculated with strain 12Đ) and 200 μL from jars inoculated with bacteriophages were serially diluted to 10-4 and spread onto TCBS agar plates supplemented with 1.5% agar. Plates were incubated at 32oC for 24 hours, and colony morphology and CFU were assessed at 3, 6, and 18 hours post-bacteriophage inoculation.
Phage genome sequencing and bioinformatics analysis
DNA extraction was performed using the Qiagen DNeasy Blood & Tissue Kit following the manufacturer’s instructions. DNA quality and quantity were assessed using a NanoDrop spectrophotometer. Sequencing libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina) and sequenced on an Illumina NextSeq 500 platform (2 x 150 bp paired-end). Raw reads were quality-controlled using FastQC v0.11.9 and de novo assembled using Unicycler (v0.4.8). All predicted open reading frames (ORFs) were compared against the NCBI non-redundant protein database using BLASTp. Whole genome alignments were performed using progressiveMauve. The genome maps were visualized using CGView.12
Statistical analysis
ANOVA (Tukey’s and Fisher’s tests) was performed using MS Excel and Minitab 16. Data are presented as means ± standard deviations (SDs) of triplicate experiments.
Results
Identification of H2S-producing bacteria
PCR electrophoresis results of the 16S rRNA gene segment showed that six samples had bands at approximately 1500 bp, consistent with the expected size. 16S rRNA gene sequencing was performed to identify the H2S-producing bacterial strain 12Đ. Phylogenetic analysis based on the 16S rRNA gene sequence placed strain 12Đ within the Desulfovibrio genus, showing close relatedness to D. vulgaris (Figure 1). Although the 16S rRNA gene sequence of strain 12Đ has not yet been submitted to GenBank, the phylogenetic analysis strongly suggests it belongs to the D. vulgaris species.
Gas production in shrimp farming water before and after inoculation with D. vulgaris
Table 1 shows that before bacterial inoculation (pH=9, salinity 15.6‰), natural shrimp pond water had negligible NH3 (0 mg/L) and H2S (0.002 mg/L), and a moderate NO3- concentration (50 mg/L). Dissolved O2 was 4 mg/L). After inoculation with D. vulgaris strain 12Đ, pH decreased (pH=8), but salinity remained at 15.6‰. H2S increased to 0.97 mg/L, NH3 increased to 5 mg/L (a dangerous level for shrimp farming), and DO decreased to 6 mg/L. NO3- levels decreased significantly.
It can be predicted that the beneficial bacteria had died off due to the significant increase of harmful bacteria in the culture jar (Figure 2A). In addition, pre-inoculation agar plating results showed that the bacterial concentration in the jar was still low, and the gases produced were not yet at a level that killed the cultured shrimp.
However, post-inoculation agar plating results showed that the concentration of D. vulgaris increased significantly, as evidenced by the black colonies on the TCBS medium (Figure 2B). This is explained by the fact that D. vulgaris bacteria produce H2S gas, which reacts with Fe3+ in TCBS medium to form ferric sulfide (FeS- black precipitate), indicating the presence of black on the colonies.2
Gas production after bacteriophage inoculation
Table 2 shows the results of gas production at 3- and 18-hours post-bacteriophage inoculation. In the control, H2S and NH3 levels remained high at both time points. At 18 hours, bacteriophage treatments, particularly ST22, φM1, φS9, φM2 significantly decreased H2S to levels safe for for shrimp (≤0.05 mg/L). NH3 levels were also significantly lower. In treatments with ST22 and φM1, DO increased to 8 mg/L, indicating effective inhibition of H2S production.
Inhibitory effects of bacteriophage on D. vulgaris
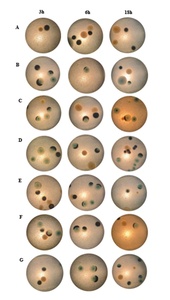
Investigating the colony morphology of D. vulgaris after bacteriophage inoculation revealed that almost all colonies lost their black color and turned blue at 3, 6, and 18 hours (Figure 3). Almost all colonies lost their black color, indicating that the bacteriophages may have inhibited the H2S-producing ability of bacteria. Specifically, bacteriophages infected and inhibited specific bacterial strain(s), thus inhibiting bacterial virulence genes, causing bacteria to lose their ability to produce toxic gases.
Three hours and 6 hours show that the colonies gradually lost their back color at the edges, indicating that the φM1 and φM2 bacteriophages were able to eliminate the H2S-producing ability of bacteria or had superior properties when combining both phages in one treatment. At 18 hours, colonies in Figures 3C and 3G show that after the bacteriophages inhibited the gas-producing ability of bacteria, the colonies still had black centers. They did not completely inhibit D. vulgaris bacteria. However, in Figures 3D and 3F, the colonies almost completely lost their black color. The specificity of bacteriophages can explain this, so the inhibitory abilities differed among treatments.
The colony counts assessment results at 3, 6, and 18 hours showed that all bacteriophages can reduce colony counts over time (Table 3). Phage strain ST22, which reduced the colony counts to 6.53 log(CFU/mL) compared to the control, showed the highest inhibitory efficiency at 3 hours, similar to phage strain S8, which reduced the colony density to 6.44 log(CFU/mL) compared to the control evidence of the highest inhibitory efficiency against D. vulgaris bacteria at 6 hours.
At 18 hours, phage strain ST22 showed the highest efficiency in reducing colony density, 6.46 log(CFU/mL) compared to the control, and the remaining treatments also reduced colony density. Thus, it can be seen that these bacteriophage strains reduced colony counts over time.
Genome analysis of bacteriophages ST231 and ST22 capable of inhibiting D. vulgaris
Phage ST231 and ST22 have complete genomes consisting of circular DNA molecules. The annotation information for ST231 and ST22 can be found on GenBank with accession numbers OP921041.1 and OQ957557.1, respectively. The genomes are visualized in Table 4. Bacteriophage ST231 has a genome size of 41,280 bp with a GC percentage of 53.9% and encodes 61 ORFs. Bacteriophage ST22 has a genome size of 45,779 bp with a GC percentage of 54.7% and encodes 62 ORFs. Both phages belong to the Caudoviricetes class. The genomes do not contain tRNA genes, lysogenic phage-related markers, toxin genes, virulence genes, or antibiotic resistance genes, making them potentially safe agents for controlling D. vulgaris infections in shrimp ponds.
Discussion
This study demonstrates the potential of bacteriophages as biological control agents against D. vulgaris in shrimp aquaculture. The reduction in H2S and NH3 levels, coupled with increased dissolved oxygen, supports previous findings on phage treatment of sulfate-reducing bacteria in wastewater.13 The loss of black coloration in D. vulgaris colonies suggests inhibition of H2S production, possibly due to phage interaction with bacterial virulence genes.14
The bacteriophages ST22 demonstrated the most promising results in the tests. Importantly, no virulence factors or antibiotic resistance genes were found in either phage genome, indicating their potential for safe use in biocontrol. Key structural genes (major capsid protein, tail protein, portal protein), lysis-related genes (endolysins and holins), and several DNA replication and metabolism genes were identified. The genome sizes and G+C content of ST231 and ST22 are typical for tailed phages. Their modular genome organization, including structural genes, lysis modules, and DNA replication genes, is consistent with their lytic lifestyle, which is advantageous for phage therapy applications.15,16 The identification of lysis-related genes, particularly endolysins, in both phages, is significant for their potential use in biocontrol.17 This host-phage relationship is crucial for effective phage therapy.18
The gradual reduction in bacterial counts suggests a sustained effect of phage treatment,19 while the enhanced efficacy of phage cocktails (φM1 and φM2 in some cases) aligns with previous research.20 However, laboratory conditions may not fully reflect complex shrimp pond ecosystems. Factors such as environmental fluctuations, other microorganisms, and potential phage resistance development must be considered for field applications.21 The genomic similarities of ST231 to Vibrio phages and ST22 to Enterobacter phages suggest the potential for controlling multiple bacterial species in shrimp ponds,22 which broadens the potential application for these phages beyond D. vulgaris control. The long-term ecological impacts of phage introduction require further investigation.23
Future research should focus on optimizing phage dosage and timing of application, as well as investigating potential synergistic effects with other biocontrol agents. Moreover, studies on the potential development of phage resistance in D. vulgaris and strategies to mitigate this risk would be valuable for sustainable long-term application of phage therapy in aquaculture.
Conclusions
This study demonstrates the potential of bacteriophages as an effective biological control agent against D. vulgaris in shrimp aquaculture. Through 16S rRNA sequencing, we successfully identified D. vulgaris in shrimp pond environments. Bacteriophage treatment resulted in significant reductions of H2S and NH3 levels, coupled with increased dissolved oxygen, indicating the phages’ ability to inhibit the gas-producing capabilities of D. vulgaris. Morphological changes in bacterial colonies post-treatment further supported this inhibition. Among the tested phages, ST22 showed the most promising results in reducing bacterial colony counts. Genomic analysis of bacteriophages ST231 and ST22 showed that they are classified under the Caudoviricetes class. The genomes do not possess tRNA genes, lysogenic phage-related markers, toxin genes, virulence genes, or antibiotic resistance genes. This suggests that these phages could be safe options for managing D. vulgaris infections in shrimp ponds. The gradual reduction in bacterial counts over time with all tested phages suggests a sustained effect, while combinations of phages showed enhanced efficacy in some cases. These results indicate that bacteriophages, particularly strain ST22, offer a promising biological alternative to antibiotics for preventing and controlling diseases caused by D. vulgaris in shrimp aquaculture. However, further research is needed to evaluate the long-term ecological impacts and efficacy of phage treatment in complex shrimp pond ecosystems before large-scale application can be recommended.
Acknowledgments
The authors are especially grateful to the Institute of Food and Biotechnology, Can Tho University, for making facilities available for this research. Van-Thanh Vo was funded by the Master, PhD Scholarship Programme of Vingroup Innovation Foundation (VINIF), code VINIF.2023.TS.111.
Authors’ Contribution
Conceptualization: Truong Thi Bich Van; Methodology: Truong Thi Bich Van, Tran Vo Minh Thu, Van-Thanh Vo; Formal analysis and investigation: Truong Thi Bich Van, Tran Vo Minh Thu, Van-Thanh Vo, Nguyen Thi Loan Anh; Writing - original draft preparation: Tran Vo Minh Thu, Van-Thanh Vo, Nguyen Thi Loan Anh; Writing - review and editing: All authors; Supervision: Truong Thi Bich Van
Competing of Interest – COPE
No competing interests were disclosed.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All are available upon reasonable request.


_and_post-inoculation_(b)_agar_plating_results_of_pond_bottom_sludge_wa.jpeg)

_and_post-inoculation_(b)_agar_plating_results_of_pond_bottom_sludge_wa.jpeg)
