INTRODUCTION
Topmouth culter (Culter alburnus) is a commonly freshwater aquaculture fish in reservoirs and ponds across various regions in China.1 This fish is highly susceptible due to its character traits of being adept at jumping, quick-tempered and easily frightened. Substantial losses occur post-netting procedures, transportation and other activities. Therefore, it’s imperative to pay special attention during these operations. Selective breeding is necessary to reduce stress response in fish, particularly in species like topmouth culter.2 Fish undergo three stages when responding to stressors (alarm reaction, resistance, and exhaustion); we mainly focus on the first stage in this study because fish in this experiment were subjected to brief and intense stress; an alarm reaction was triggered during which stress hormones were released and energy was required.3 Other slow and long-term stress, such as high-density farming or disease stress, will trigger the second or third stage of response. A deeper understanding of the biological mechanisms underlying stress responses in relevant tissues can help enhance the welfare and production efficiency of fish.4
Transcriptome profiling is an effective method to assess differential pathways and gene expression involved in the altered culture environment of aquatic animals.5 In the transcriptome of the head kidney and mixed tissue of fish, many immune-related genes and signaling pathways have been identified.6,7 Yet, the tissue-specific stress-related genes and pathways in topmouth culter remain unidentified. Transcriptome analysis had been carried out under different stress conditions in the brain and kidney of rainbow trout (Oncorhynchus mykiss),8 as well as in the liver, muscle and kidney of red cusk-eel (Genypterus chilensis),3,9 many genes and pathways were identified in these studies, providing useful information for fish breeding and scientific research. Results obtained from rainbow trout and red cusk-eel demonstrated that RNA-seq will be a valuable tool for examining tissue-specific stress responses in topmouth culter.
The Gene Ontology is becoming increasingly utilized for the analysis of gene expression profiles in various treatments or tissues following RNA-seq.10 The response to stimulus(GO:0050896), the response to stress (GO:0006950) and the response to hypoxia (GO:0001666) were three significant stress-related GOs.11–13 In the liver of rainbow trout, genes such as haptoglobin and complement factor H-related protein 1 were annotated and significantly altered in the biological process category “response to stimulus” after experiencing a handling stressor.14 Identifying and analyzing tissue-specific differential genes across the three GO terms mentioned above will yield valuable insights into the future functional studies of candidate genes involved in stress response.
Hence, we conducted de novo transcriptome sequencing of the kidney, muscle, and liver in the current study. Unlike previous extensive studies, this research separately and significantly demonstrated various skeletal muscle-related responses in muscle tissue, fat metabolism in the liver, and immune response-related pathways in the kidneys. These findings offer novel insights into stress research. Furthermore, gene ontology enrichment analysis identified 18 genes significantly associated with stress response. Their expression was verified by qPCR under various stress conditions (chasing and netting out of water). These findings will offer additional molecular markers for the selective breeding of topmouth culter. Leveraging these molecular markers can enhance the breeding traits, ultimately fostering the swift and healthy growth of the topmouth culter industry.
MATERIALS AND METHODS
Biological samples and RNA preparation
10 three-year-old female topmouth culter (body weight 671.21 ± 75.25 g and body length 37.78 ± 1.57 cm) were netted from Taihu Lake (Zhejiang Province, China). Fish were anesthetized in 100 mg/L tricaine methane sulfonate (MS-222, Sigma, St.Louis, MO) for ten minutes, then kidney, liver and muscle tissues of each fish were quickly removed, snap-frozen and stored at -80℃ for RNA-seq. Total RNA was extracted using TRIzol Reagent (Invitrogen, USA). After DNase treatment, the quality of total RNA was determined using a Nanodrop 1000 spectrophotometer (Thermo, USA) and gel electrophoresis. High-quality mRNA from the kidneys, liver, and muscle of 10 individuals was pooled in equal amounts for sequencing.
Library construction and High-throughput sequencing
cDNAs were synthesized using Illumina TruSeq RNA Sample Preparation Kit, which contains Oligo (dT). After being suspended in the fragmentation buffer, mRNA was randomly broken into small pieces, and random hexamers were then used for cDNA synthesis. After being purified by QIAquick PCR Purification Kit (Qiagen, GER), the cDNA was amplified, and libraries were constructed. Then libraries with an average length of ~200bp were used for sequencing on an Illumina HiSeq 2000 to generate 100bp paired-end reads.
De novo assembly of sequencing reads and sequence analysis
Raw data generated from Illumina sequencing were trimmed by removing adapter sequences, low-quality bases, reads with unknown base calls more than 5% and reads with length less than 20 bp. Transcripts were assembled by using Trinity software.15 To acquire non-redundant unigenes, TGICL software was then used to sequence splice and remove redundancy.16
BLASTx searches were performed to align all assembled transcripts to the non-redundant, Swiss-Prot, COG, GO and KEGG database for functional annotation (E-values<1e-5 and identities>30%). Gene identity was assigned to each protein sequence based on the best BLAST hit. Gene ontology analysis was conducted on assembled transcriptome sequences by using Blast2GO program, a software package that retrieves GO terms.17 After obtaining the GO annotation for every topmouth culter unigene, WEGO software was further carried out to predict the functions of unigenes. And Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was analyzed using KEGG Automatic Annotation Server (KASS) with default parameters.
Identification of different expression genes in three stress-related GOs
Tissue-specific gene expression was carried out by FPKM (Fragments Per kb per Million fragments) method in topmouth culter kidney, liver and muscle transcriptome. FDR (False Discovery Rate) was used to correct for multiple comparisons. Unigenes with fold changes >2 and FDR<0.001 were defined as differential expression genes. Screening of DEGs was followed by GO functional enrichment analysis and tissue-specific gene were then identified in three stress-relative GO, response to stimulus (GO:0050896), response to stress (GO:0006950) and response to hypoxia (GO:0001666). Using phyper algorithm test, significantly enriched metabolic pathways or signal transduction pathways in DEGs were identified between tissues.
Putative SNP and SSR identification
The SNP (Single Nucleotide Polymorphism) detection was performed using the SAMtools tool kit. A putative SNP site should satisfy the following conditions: (1) both alleles were detected from the contigs and unigenes; (2) the read coverage was at least 2 reads; (3) the minor allele frequency was at least 20%. SSR (Simple Sequence Repeats) of topmouth culter were identified from the unique sequences using the MISA program. A minimum number of repetitions of dinucleotide was set at six, and trinucleotide, tetranucleotide, pentanucleotide, and hexanucleotide were set at five. Two SSRs separated by more than 100 bp were considered to be two SSRs.
Fish management, acute stress exposure and sample collection
One hundred and eighty 16-month old topmouth culter with an average weight of 150.3 ± 21.3 g, average length of 230.3 ± 15.2 mm were collected from the comprehensive experimental base of Zhejiang Institute of Freshwater Fisheries. Fish were distributed evenly into 18 tanks (10 fish/tank, 300L PVE tanks, aeration) and then acclimated for 10 days under controlled conditions (temperature 22.5 ± 3.2°C; dissolved oxygen 7.0 ± 0.6 mg/L; 12 h light: 12 h dark cycle). Commercial pellets (Tongwei, China) were fed daily with 1% body weight of fish during acclimation. Three groups (control group, treatment group and recovery group) were randomly divided with 60 fish in each group. Before treatment, 12 fish were sampled from the control group at 0 points and then stayed undisturbed during the experiment. Fish from the handling group and recovery group were subjected to two repeated handling stresses, including chasing for 2 min and netting out of water for 30 sec according to the method of Castanheira18 and Aedo9 with some modifications. Fish in the treatment group were killed immediately after handling, while fish in the recovery group were quickly sampled at 24 hours post-stress. All fish were ethically euthanized with 100 mg/L tricaine methane sulphonate (MS-222, Sigma), and blood, kidney, liver, and white muscle samples were sterile collected and then frozen for further processing (n = 12 per group). Plasma cortisol was determined with ELISA using corresponding kits (H094, Nanjing Jiancheng Bioengineering Institute).
qPCR expression analysis
Kidney, liver, and muscle RNAs from three groups were extracted and reverse transcribed as described above. 18 Primers for tissue-specific genes and internal control gene β-actin19 were designed by the software Primer Premier 6 and listed in Table 1. Analyses were carried out using SYBR green (Takara, Japan) and a Roche 480 light cycler system. Each qPCR was carried out in triplicate. The qPCR program was an initial cycle activation at 95°C for 2 min, 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. 2−ΔΔCT method was then used to analyze the average fold change of target genes.
Statistical analysis
Data were analyzed using SPSS 13.0 software (one-way analysis of variance followed by Duncan’s multiple range test) and shown as standard error (S.E.) of the means. Results were considered statistically significant when P < 0.05.
RESULTS
SEQUENCE ANALYSIS AND ASSEMBLY
To obtain an overview of topmouth culter tissue-specific gene profile, cDNAs from kidney, liver and muscle tissues of ten female fish were generated and sequenced using illumina sequencing platform. A total of 28,781,812 raw reads from topmouth culter kidney, 28,022,888 raw reads from liver, and 29,041,506 raw reads from muscle tissue were generated, respectively. These reads have been deposited in NCBI Sequence Read Archive database with the accession number SRR2182152 for kidney tissues, SRR2179946 for liver, and SRR2182178 for muscle. After the removal of adapters, short and low-quality sequences, a total of 25,795,994, 25,745,310, and 27,322,110 clean reads were subjected to downstream analysis (Table 2). By assemble analysis, the lengths of CDSs ranged from 102 bp to 21,594 bp with an average length of 423 bp (Figure 1). Kidney tissue had the highest contig count, 193,502 ESTs (117,760 contigs averaging 332 bp and 75,742 unigenes averaging 523bp) were obtained from the kidney transcriptome. additionally, the lowest contig count was observed in liver tissue, with only 91,765 ESTs (57,607 contigs averaging 308bp and 34,158 singlets averaging 484bp). There were 149,915 ESTs (92,707 contigs averaging 296bp and 57, 208 singlets averaging 460bp) from the muscle transcriptome (Table 3). The length distribution of contigs and unigenes is presented in Figure 1.
ANNOTATION AND FUNCTION ANALYSIS
In total, about 79.1%, 49.3%, 43.5 % and 36.2% of the topmouth culter nonredundant unigenes had significant matches with sequences in NT, NR, Swiss-Prot and KEGG databases, respectively. The top ten KEGG pathways of unigenes in three tissues are listed in Table 4. The most represented pathways were the “Metabolic pathways” and the “Regulation of actin cytoskeleton”. Some pathways related to immune or signal transduction were also identified, such as the “Pathways in cancer” and “Focal adhesion,” “Dilated cardiomyopathy,” and “Calcium signaling pathway.”
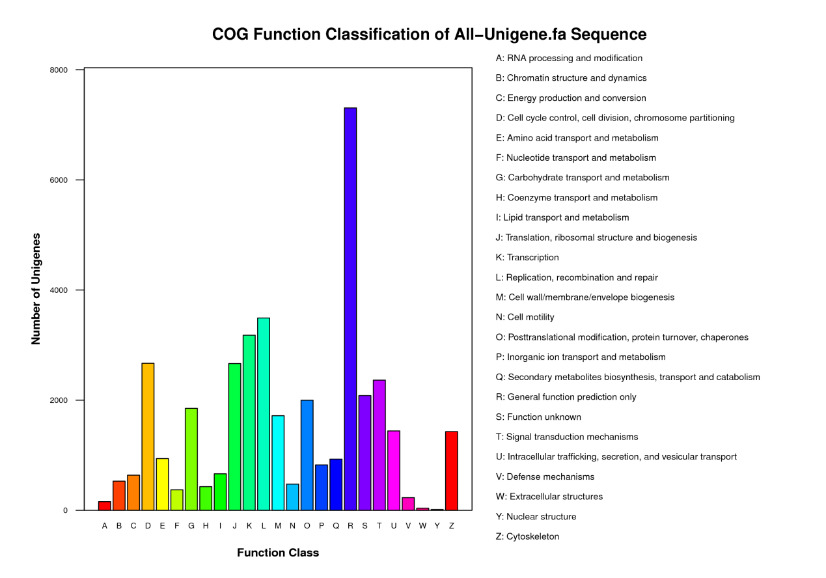
To predict possible functions of the acquired transcriptome, 16,411unigenes were classified into 25 COG categories by aligning to the COG database (Figure 2). Among these, the largest functional classes were the “general function prediction” (R category, 19.01%), followed by “replication, recombination and repair” (L category, 9.09%), “transcription” (K category, 8.27%), “cell cycle control, cell division, chromosome partitioning” (D category, 6.94%), “translation, ribosomal structure and biogenesis” (J category, 6.93%) and “signal transduction mechanisms” (T category, 6.15%). Only 5.42% unigenes were assigned to “function unknown” (S category).
All unigenes were assigned to different GO functional groups according to their functions. There was no significant difference in GO classification among kidney, liver and muscle transcriptome (Figure 3). In the cellular components category, cell (about 21%) and cell part (about 21%) followed by organelles (about 15%) and membrane (about 10%) were represented. Binding (about 47%) and catalytic activity (about 27%) predominated the molecular function category. In the biological process, cellular process (about 17%), single-organism process (about 13%), metabolic process (about 13%), and biological regulation (about 11%) were four primary subcategories.
SSR AND SNP ANALYSIS
A total of 16269 SSRs were identified from kidney, liver and muscle tissues using the next-generation sequencing. The most common repeat motifs were mono-nucleotide repeats (44.5%), other repeat motifs: Di-nucleotide repeats (35.1%), Tri-nucleotide repeats (16.4%) and tetra/penta / hexanucleotide repeats (4.0%) were also detected. We found that AC/GT and ATC/ATG repeat motifs were the most abundant motifs in Di- and tri- nucleotides, respectively (Figure 4). Additionally, we also identified 23,854, 10,313 and 11,436 predicted SNPs in kidney, liver and muscle transcriptome respectively. The proportions of different transition types in three tissues were almost equal (about 50% for A/G and 50% for C/T), the proportions of four transversion types (about 30% for A/T, 27%A/C, 25% for G/T and 18% for C/G) were also very similar (Figure 4).
TISSUE-SPECIFIC GENE EXPRESSION IN THREE TRANSCRIPTOMES
Comparative analysis results showed that 10,099 unigenes were co-expressed in three tissues, while 3158, 409 and 1952 unigenes were expressed only in kidney, liver and muscle transcriptome (Figure 5A). In addition, comparison analysis of DEGs using the criteria FDR ≤0.001 and |log2Ratio| ≥1 showed that a total of 27025, 16322 and 25534 unigenes were differentially expressed in liver vs kidney, liver vs muscle and kidney vs muscle groups of topmouth culter transcriptome, numbers of unigenes which was down-regulated and up-regulated in each group were showed in Figure 5B. For tissue-specific GO analysis, 18, 4, and 11 genes were unique expressed in the kidney, liver, and muscle when compared with the other two tissues in response to stimulus GO (GO:0050896), the number of gene were 15,4 and 12 genes in response to stress GO (GO:0006950) and 10,4 and 5 genes in response to hypoxia GO (GO:0001666), respectively (Table 5). Detailed information of 6 genes of each GO above is listed in Table 6.
CHANGE OF PLASMA CORTISOL LEVEL IN RESPONSE TO HANDLING STRESS
Levels of cortisol in treatment and recovery groups changed significantly after stress administration (Figure 6). There was around 2.7- and 1.8-fold increase than that of the control group (P < 0.05).
CHANGE OF TISSUE-SPECIFIC GENE IN KIDNEY, LIVER AND MUSCLE TISSUE IN RESPONSE TO HANDLING STRESS
In response to stimulus GO, the expression of the LCP2 gene in the kidney and TNFRSF5 gene in the liver were significantly increased, while the expression of COL11A2 and MYH10 gene in the muscle was significantly lower than those in the control group (P < 0.05). In the recovery group, except for the significant increase of COL11A2 gene, the other five genes (LCP2, P-selectin, CYP4V2, TNFRSF5 and MYH10) were all lower than those in the control and treatment group, significant (P < 0.05) decrease was detected in four genes (LCP2, P-selectin, CYP4V2 and MYH10) (Figure 7).
In response to stress GO, except for the Sep 1 gene, the expression trend of the other five genes decreased gradually after administration. Significant changes were noted in the Serping 1 gene in the liver, Myogenin 2, and Troponin C gene in muscle tissues in the treatment group. AhR1, Renin, ADRA1a, Myogenin 2 and Troponin C genes decreased significantly (P < 0.05) compared with the control group in recovery group (Figure 8).
In response to hypoxia GO, in the treatment group, the PTK2b gene in the kidney and the FABP1 gene in the liver increased significantly. Meanwhile, RYR1b and CAPN2 genes in the muscle showed decreased significantly (P < 0.05). In addition, in the recovery group, except for the significant increase of the IGFBP1 gene in liver tissue (P < 0.01), expression of the other five genes (CCR, PTK2b, FABP1, RYR1B, and CAPN2) were significantly (P < 0.05) lower than those in the control group (Figure 9).
DISCUSSION
TRANSCRIPTOME ASSEMBLY OF THREE TISSUES IN TOPMOUTH CULTER
According to previous studies, sequencing depth and the reads quality filtering in this study were optimal for tissue-specific transcriptome assembly.20 However, there were still 20.9% sequences without any BLASTx hits, 5.42% unigenes were assigned to “function unknown” in COG analysis, and 64.38% unigenes were not annotated into KEGG databases. The efficiency of annotation and assignment in this study was comparable to transcriptome sequencing studies conducted in the spotted sea bass (Lateolabrax maculatus)21 and miiuy croaker (Miichthys miiuy).22 These results speculated that relatively low annotation efficiency might be attributed to incomplete assembly and lack of genomic information for these fish.21 Furthermore, SSRs and SNPs have been extensively utilized in parentage analysis and population genetics studies across various fish species. We analyzed the SNPs within the TGFβ-smads pathway genes (including Smad4, TGFβ and inhibin βB) in three generations of gynogenetic topmouth culter.23 While some SSR were identified in the topmouth culter, the number and detection method were limited.24,25 Therefore, the data obtained from this study can be regarded as potential candidate loci for analyzing population diversity and paternity in molecular-assisted breeding of topmouth culter.
TISSUE-SPECIFIC GENE ONTOLOGY ANALYSIS
Gene ontology provides a powerful means for searching functionally related genes. Only 12 unique genes were identified in liver tissue in this study. This may be attributed to the fact that the number of assembled contigs and unigenes in the kidney and muscle transcriptomes was greater than that in the liver tissue. It also indicated that there might be some more complex stress reactions in the kidney and muscle tissues than those in the liver tissue. Hence, it is necessary to study these genetic response mechanisms in specific tissue after stress in topmouth culter.
EFFECT OF HANDING STIMULATION ON CORTISOL OF TOPMOUTH CULTER
When fish is stimulated by stress factors, the hypothalamic-pituitary-adrenal (HPA) axis responds quickly, leading to the synthesis and release of cortisol hormones in the head kidney cells. Thus, the cortisol concentration in the blood can typically serve as a marker to assess the stress levels in fish.26 In our study, a series of persistent and intense irritants were applied to the treatment and recovery groups of topmouth culter, revealing a pattern of initial cortisol increase followed by a decrease. Notably, the cortisol levels of European sea bass (Perca fluviatilis) and spotted catfish (Ictalurus punctatus) also rose significantly during post-stress,27,28 aligning with our experimental findings. Elevated cortisol levels might trigger alterations in numerous subsequent physiological processes. In red cusk-eel, handling stress induced increasing in cortisol levels and significant changes in skeletal muscle gene expression. These suggested that cortisol was a potent inducer of skeletal muscle atrophy in fish myotubes.9 However, the mechanism by which cortisol contributes to the stress response in the topmouth culter still requires further validation.
EFFECT OF HANDING STIMULATION ON STRESS-RESPONSE GENE IN THREE GOS
In response to stimulus GO, which is the process begins with the detection of the stimulus and ends with a change in state or activity. After 24 hours of treatment in this study, significant down-regulation of three genes (P-selectin, CYP4V2 and MYH10) in the recovery group was observed. Specifically, P-selectin was found to increase leukocyte recruitment and leukocyte rolling in both rabbits and rats.29,30 Furthermore, in diabetic human kidneys, P-selectin had been shown to increase interstitial capillaries and glomerulus, which was involved in the development of inflammatory cytokines.31 These results suggested that P-selectin may also play a role in regulating kidney function during the recovery period after exposure to external stimuli in fish. In the CYP4 family, it was found that CYP4V2 performs oxidation-reduction reactions in fatty acid metabolism in both human and chicken hepatocytes.32,33 After recombinant CYP4V2 protein metabolized with DHA, ω and ω-1 hydroxy metabolites were formed.34 In this study, we observed a significant suppression of CYP4V2 expression during the recovery period, along with a concurrent decrease in FABP1 gene expression in the liver. It suggests that fatty acid metabolism process might be disrupted in the liver of topmouth culter during the recovery period. MYH10 has been previously linked to tumor pathogenesis, promoting cell proliferation, invasion, and metastasis,35 as well as centriole migration in the biogenesis of primary cilia.36 The inactivation of MYH10 might lead to embryonic cytokinetic failure in cardiac myocytes.37 In summary, these three genes (P-selectin, CYP4V2 and MYH10) are closely associated with diseases and immune function, indicating that the immune system of topmouth culter had been activated and might lead to the subsequent development of related diseases.
Response to stimulus GO is the parent term of response to stress GO, the synonyms of response to stress is any process that results in abiotic or biotic stress. Notably, significant changes were observed in the Serping 1 gene in the liver and the Myogenin 2 and Troponin C genes in muscle tissues of the treatment group. For the Serping 1 gene, following the stimulation by silver nanoparticles toxicity, the Serping gene in rainbow trout was found to be involved in the regulation of the complement C1 system. Concurrently, decreases in Serping gene expressions in liver tissue were observed correlating with an increase in lipid peroxidation (LPO).38 While liver LPO indicators were not monitored in this study, changes in cortisol content were detected. After 96 hours of low-water stress treatment, the levels of LPO and cortisol in the liver of the chinook salmon (Oncorhynchus tshawytscha) increased synchronously. It was hypothesized that stimulation at low water levels, as observed in this study, might elicit a response in the lipid peroxide metabolism process of the fish liver.39 Previous research suggested that the Serping 1 gene might regulate external stimuli through immunity and lipid peroxidation in topmouth culter liver. Consistent with the findings of this experiment, the expression of the troponin C gene was significantly suppressed in rainbow trout after 5 days of handling stress.8 In Zebrafish, the downregulation of troponin might contribute to muscular dysfunction.40 Troponin C, troponin T and troponin I combine to form the troponin complex, which regulates the interactions between myosin and actin filaments in muscle tissue.41 Myogenin is a member of the myogenic regulatory factors family.42 The higher expression level of myogenin in slow-twitch muscle suggested a significant role in continuous swimming in Pseudocaranx dentex. Additionally, differentiated expression patterns of troponin genes were observed between slow-twitch and fast-twitch muscle.43 In this study, it was observed that during the stress-handling process, the muscles of topmouth culter were vigorously exercised. However, during the recovery period, the swimming speed of the fish decreased, even falling below their normal speed. This could be the primary reason for the significant reduction in the expression of genes related to force generation and energy release, such as troponin C and Myogenin 2 gene.44
For response to hypoxia GO: Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%. At the cellular level, hypoxia might lead to the production of reactive oxygen species, reactive nitrogen species and increased concentrations of nitric oxide synthase.45 PTK2 gene, also known as tyrosine kinase 2 (TYK2) gene, is a member of the tyrosine kinase family, it plays a crucial role in regulating cell growth, development, innate, and adaptive immunity.46 It exhibits a high degree of cell type and tissue specificity in its expression. Activation of PTK2B is triggered by phosphorylation on tyrosine residues in response to a range of stimuli.47 Notably, the TYK2 gene in the head kidney of golden pompano (Trachinotus ovatus) showed inducible expression post LPS, poly I:C and Vibrio alginolyticus stimulation, highlighting its significance in the response to pathogen invasion.47 Fatty acid–binding protein1 (FABP1) is a liver-specific FABP. Over-expression of FABP1 led to a significant increase in hepatocyte fatty acid uptake.48 Notably, elevated expression of FABP1 in the liver of javelin goby (Synechogobius hasta) and largescale shoveljaw fish (Onychostoma macrolepis) suggested its role in energy metabolism following fasting in these species.49,50 During the recovery phase, the appetite of topmouth culter was initially suppressed, they resumed autonomous feeding 24 hours post-stress. Consequently, we examined the expression of FABP1 and observed its suppressed expression. Research demonstrated that the energy metabolism in fish returns to the pre-treatment level at the molecular level after 24 hours. Both RYR1b and CAPN2 are Ca2+-dependent receptor/protease, playing roles in distinct cellular processes within fish muscle. In particular, training programs led to a significant increase in the detection of Rryanodine receptor gene in brown trout (Salmo trutta) and whitefish (Coregonus lavaretus) muscle. Throughout these processes, the Ca2+ regulatory ability had been significantly negatively impacted.51,52 To date, no studies have examined the impact of hypoxia on CAPN2 gene expression in fish. Under distinct nutritional conditions (starvation/ refeeding) in rainbow trout and Atlantic halibut, it had been demonstrated that CAPN2 gene expression was involved in energy metabolism processes in muscles.53 Consequently, hypoxia may significantly affect ion channels in the skeletal muscle membrane by altering the expression levels of related genes.54
However, it is worth noting that tissue-specific expression was not the primary focus, leading to the potential loss of valuable information in this study. In addition, it has been observed that certain genes exhibit varied expression patterns after stress when compared to other fish species. This variation might be influenced by the species of fish, duration and intensity of stress. For the complex physiological process of emergency stress, a mere analysis of gene expression and hormone levels might be insufficient. Hence, further studies will focus on the protein level to provide a more comprehensive understanding of the precise physiological responses of topmouth culter to external stress.
CONCLUSION
Our study aims to compare the acute stress responses of the kidneys, liver and muscle of topmouth culter, in order to analyze the internal mechanisms of irascible and easily frightened. Contrasting the responses of the three tissues, it was observed that the kidney and muscle tissue showed a stronger reaction to challenges than the liver tissue. Through specific GO restrictions, we identified only a few specific genes (no more than 20) with unique functions. All of these genes changed with handling operations and cortisol levels. This study preliminarily confirmed that these genes might serve as reliable analytical indicators for studying stress in topmouth culter. This study provided new focus for breeding of topmouth culter industry, and it also offered a reference for measuring stress in diverse conditions in other fish.
ACKNOWLEDGMENTS
This study financially supported by Key Scientific and Technological Grant of Zhejiang for Breeding New Agricultural Varieties (2021C02069-3).
AUTHORS’ CONTRIBUTION
Conceptualization: Meili Chi (Lead). Investigation: Meili Chi (Equal), Shun Cheng (Equal), Jianbo Zheng (Equal), Shili Liu (Equal), Wenping Jiang (Equal). Formal Analysis: Meili Chi (Equal), Wenping Jiang (Equal), Fei Li (Equal). Writing – original draft: Meili Chi (Lead). Supervision: Wenping Jiang (Equal), Fei Li (Equal). Writing – review & editing: Wenping Jiang (Equal), Fei Li (Equal).
COMPETING OF INTEREST
No competing interests were disclosed. The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
ETHICAL CONDUCT APPROVAL
The sample collection and experiments in the study were in compliance with the Animal Ethics Committee of Zhejiang Institute of Freshwater Fisheries (Animal Ethics no. 1067, March 6, 2019).
INFORMED CONSENT STATEMENT
All authors and institutions have confirmed this manuscript for publication.
DATA AVAILABILITY STATEMENT
All data generated or used during the study appear in the submitted article.




_and_six_types_of_snp_(b)_in_kidney__liver_and_muscle_t.png)
_and_different_expression_gene_(deg)(b)_among_thes.tiff)






_and_six_types_of_snp_(b)_in_kidney__liver_and_muscle_t.png)
_and_different_expression_gene_(deg)(b)_among_thes.tiff)



