Introduction
Weissella species are typically not associated with disease in fish. However, it has been recently found that Weissellosis in farmed rainbow trout (Oncorhynchus mykiss) is induced by a Gram-positive lactic-acid bacteria, Weissella ceti.1 This kind of infectious pathogen has been detected successively in Asia, South America, and North America.2 Furthermore, it can lead to higher mortality rates from 60% to 80% in rainbow trout, especially in those weighing 0.25 - 1 kg, resulting in considerable economic losses for local aquaculture industries.3 The clinical manifestations in infected rainbow trout include hemolysis in various tissues, anorexia, lethargy, darkening of the skin, ascites, exophthalmia, corneal opacity and even occasional corneal rupture.4–7 It is important to note that these symptoms are not entirely pathognomonic for Weissellosis. Thus, there is a pressing need for the effective detection of W. ceti to prevent their occurrence and spread.
So far, the identification of W. ceti has been facilitated through biochemical, physiological, phenotypic characterizations, along with bacteriological cultures and molecular biological techniques, particularly those based on PCR.1,8–11 However, these methods have certain limitations, for example, the requirement for extended timeframes, costly and sophisticated equipment, and specialized personnel. Such constraints render these methods impractical for point-of-care testing (POCT) diagnosis in the aquaculture industry. Therefore, it is necessary to develop a rapid and convenient diagnostic tool to identify W. ceti, especially at the early stage of the infection.
Loop-mediated isothermal amplification (LAMP), a nucleic acid amplification technique, was initially developed by Notomi.12 Subsequently, the amplification process was accelerated by the additional loop primers.13 Two or three primer pairs (inner, outer, and loop primers) for the LAMP reaction can quickly recognize six or eight specific regions on the target gene or sequence and strongly improve the specificity. This amplification is performed using strand displacement DNA polymerase under isothermal conditions, resulting in the generation of a multitude of amplicons with characteristic multiple loops.14 There exists a great diversity of methods to detect LAMP reaction products. For example, gene amplification can be observed by the naked eye either in the form of turbidity or a color change without any additional process. Such a detection method is particularly amenable to point-of-care testing (POCT) diagnosis. Moreover, the LAMP reaction does not require a thermal cycler, so it can be accomplished only by depending on a simple heater without expensive or advanced laboratory equipment. Additionally, this method can avoid the risk of carryover contamination by a one-step, one tube visualization detection system.15
Because LAMP has rapid, convenient, specific, and sensitive detection capabilities, it can be accomplished in a simple laboratory environment, which is particularly suitable for POCT.16 This method has been extensively applied for the detection of all kinds of organisms, such as viruses,17 bacteria,18–20 fungi21 and parasites.22 So far, there is no reference to the detection and identification of W. ceti by LAMP.
The hemolysin A gene, which encodes the cytotoxic factor hemolysin, had been applied to the identification of other pathogenic bacteria.23,24 In the present study, colorimetric LAMP targeting the hemolysin A gene was utilized for W. ceti. This method simplifies the process and minimizes the risk of carryover contamination. The colorimetric LAMP by color change offers a rapid and simple means of identifying W. ceti compared to conventional PCR. It holds potential for application in the detection of pathogens as a rapid and highly sensitive POCT to provide reliable references to aid in the prevention of pathogen dissemination.
Materials and Methods
Primer information
For this study, the hemolysin A gene of W. ceti (sequenced by Novogene Co., Ltd*)* was targeted for LAMP. A primer set was designed according to eight different regions of the target sequence using LAMP Primer Explorer V5 (http://primerexplorer.jp/v5e/ index.html).
The primer set contained a pair of outer primers (F3 and B3), a pair of inner primers (FIP primer included F1c and F2, BIP primer included B1c and B2) and a pair of loop primers (LF and LB) (Fig. 1). The outer primers were also used as primers in PCR amplification. The primer set was synthesized by Tsingke Biotechnology Co., Ltd.
Bacterial strains
In order to ascertain the specificity of LAMP, 7 bacterial strains containing one W. ceti strain WS105, Aeromonas salmonicida, Aeromonas veronii, Streptococcus iniae, Citrobacter gillenii, Aeromonas hydrophila and Hafnia alvei were employed. All strains were sourced from the strain bank in our own laboratory. The W. ceti strain was grown in liquid Man Rogossa Sharpe (MRS). S. iniae was cultured in liquid Trypticase Soy Broth. The other 5 strains were cultivated in liquid Luria–Bertani broth. All strains were maintained at 28 ℃ for approximately 16 hr with constant shaking (180 rpm).
Subsequently, bacterial genome DNA was extracted from each strain, following the manufacturer’s illustrations of bacteria DNA isolation Mini kit (Nanjing Vezyme Biotech Co., Ltd, China). DNA extracted were tested by a DS-11 spectrophotometer (DeNovix, United States) for the concentration and quality. The qualified DNA was stored at -20°C until used.
PCR and LAMP reaction
The 25 μl PCR reaction mixtures contained 12.5 μl of 2 × Taq PCR Master Mix (TransGen Biotech Co., Ltd, Beijing, China), 1 μl of each primer, 1 μl genomic DNA template, and 9.5 μl double-distilled water (ddH2O). The PCR reaction was accomplished in a BIO-RAD T100 Thermal Cycler PCR with the following steps: a pre-denaturation at 92 ˚C for 3 min, and after that, 30 cycles including denaturation at 92 ˚C for 1 min, annealing at 61 ˚C for 1 min, extension at 72 ˚C for 1 min, and extension at 72 ˚C for 5 min as the final step.
The LAMP reaction mixtures contained 12.5 μl WarmStart Colorimetric LAMP 2 × Master Mix with UDG (New England Biolabs), 2.5 μl 10 × primer mixtures (2 μM F3 and B3, 16 μM FIP and BIP, 4 μM LF and LB) and 1 μl genomic DNA template, then adding ddH2O up to 25 μl, and incubated at 63 °C for 30 min. Except for the reaction mixtures, the DNA template was replaced by ddH2O as control groups. Both the PCR and LAMP products were electrophoresed in a 2% agarose gel at 160 V for 25 min, then analyzed and taken photos under a UV transilluminator. The products of LAMP can also be distinguished with the naked eye based on the color change.
Temperature and time of the LAMP reaction
To adopt the optimum temperature of LAMP, the reactions were conducted across a range from 60 to 65 ℃ with an interval of 1℃. In addition to temperature, the reaction duration was also optimized by setting various time points at 10, 20, 30, 40, 50, and 60 min.
Specificity and Sensitivity Analysis
Seven strains were used to confirm the specificity of LAMP. The genomic DNA extracted from seven strains (A. salmonicida, A. veronii, S. iniae, C. gillenii, A. hydrophila, H. alvei and W.ceti) were utilized as the templates for LAMP.
The extracted genomic DNA of W.ceti was prepared into a series of 10-fold dilutions with ddH2O, ranging from 5.9 100 ng/μl to 5.9 10-7 ng/μl. Utilizing these dilutions as templates, the sensitivity of PCR and LAMP was evaluated based on the minimum concentration of the genomic DNA.
Detection of simulated clinical samples
Rainbow trout with an average weight of 10 ± 2.0 g were gained from an aquaculture farm (Shandong, China). They were fed in aerated tanks at 18.0 ± 1.0 ℃ at the laboratory. Thirty rainbow trout were divided randomly into experimental and control groups. Fish of the experimental groups were injected intraperitoneally with 40 μl bacterial solution with a concentration of 1.6 108 CFU/ml, and the control groups were injected with 40 μl phosphate-buffered saline (PBS). After 24 hr, liver, spleen, and kidney tissues were sampled in a sterile environment. The tissues collected were individually mixed with 200 μl PBS and then homogenized thoroughly. Subsequently, the 10-fold dilutions of homogenate were handled in ddH2O. 100 μl from each dilution was evenly spread on MRS agar to determine the concentration of bacteria incubated in 48 hr at 28 ℃. In parallel, 100 μl from each dilution was also boiled at 95 ℃ for 5 min, followed by centrifugation at 12,000 g for 5 min. The supernatant isolated was used as DNA templates for LAMP detection.
All detection or reaction processes were included at least in triplicate for each sample.
Results
Option of temperature and time
To adopt the optimum temperature of LAMP, electrophoresis of the amplification products across a temperature gradient from 60 to 65 °C was conducted. This analysis showed typical DNA ladder-like bands and the brighter ones were from 63 to 65 ℃ (Fig. 2). In this study, 63 ℃ was adopted for the subsequent LAMP of W. ceti in view of energy conservation.
At 63 ℃, six different time points from 10 to 60 min with 10-minute intervals were set to complete LAMP. The products of LAMP were electrophoresed in a 2% agarose gel, which revealed ladder-like bands at each of the time points (Fig. 3). Based on the electrophoretic results, a reaction duration of 30 min was accepted for the subsequent LAMP.
Specificity Analysis
7 strains were selected to detect the specificity of LAMP, only the W. ceti strain showed the ladder-like bands, while the remaining six strains and the control group with ddH2O were negative (Fig. 4a). Furthermore, only the W. ceti strain was yellow (Fig. 4b), which was consistent with the results of electrophoresis. Notably, LAMP did not yield any false-positive or false-negative results, which confirmed its high specificity.
Sensitivity Analysis
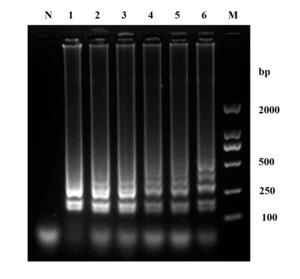
To ascertain the sensitivity of LAMP for W. ceti, a series of 10-fold dilutions with genomic DNA ranging from concentration of 5.9 100 to 5.9 10–7 ng/μl were evaluated. The minimum concentration of LAMP was 5.9 10–6 ng/μl (Fig. 5a). The minimum concentration for the PCR with outer primers were 5.9 10–4 ng/μl (Fig. 5b). Compared to traditional PCR, LAMP was 100 times more sensitive than PCR.
Applicability of LAMP
To verify the effectiveness of LAMP in clinical samples, supernatants isolated from the liver, spleen, and kidney tissues of both experimental and control groups were used to accomplish the detection. The results revealed that W. ceti could be detected in all tissues collected by the color change of LAMP reaction mixtures, and control groups were negative (Fig. 6). LAMP for W. ceti in liver, spleen, and kidney tissues demonstrated a minimum concentration of 18, 2.1, 4.4 CFU/reaction, respectively.
Discussion
One key advantage of developing an efficient molecular technique is the ability to target and select a specific genetic sequence. The specificity of the LAMP method is particularly high because the primer sets must recognize completely six different regions on the target sequence.12,25–27 The amplification reaction will not proceed if either the outer or inner primers do not recognize the target sequence. In the current study, the results indicated the specificity in LAMP for W. ceti subjected to 7 strains, without false-positive amplification observed. LAMP with higher specificity has also been confirmed in a wide variety of microorganisms.28–31
In addition to the specificity of amplification, sensitive detection is equally crucial for disease diagnosis because bacterial concentrations are often low, especially at the primary stage of the infection. Under normal conditions, the sensitivity of LAMP is 10-100 times higher than traditional PCR, but it might also be affected by various factors.32 For example, loop primers can accelerate the efficiency of target sequence amplification to enhance the sensitivity of LAMP.13 The sensitivity of LAMP was also dependent on the means used to detect the amplification products. To date, the products of LAMP may be monitored by observation of precipitate or turbidity,33 DNA-binding dyes,34 colorimetric indicators,35 gel electrophoresis36 and so on. Among these, colorimetric indicators by visual inspection offer a more convenient means compared to alternative methodologies. The most convenient colorimetric indicators mainly were calcein, hydroxy naphthol (HNB), and phenol red (PR), which have been shown to produce a color change because of DNA amplification and to mitigate more effectively the risk of carryover contamination. These indicators facilitate visual detection in a single tube and are well-suited for POCT diagnosis. Though the color change indicative of pH-sensitive indicators can be interfered by reaction mixtures, the disadvantage of colorimetric detection with PR can be mitigated under standardized LAMP conditions with minimal buffering.37,38 In the present study, LAMP with PR demonstrated high sensitivity with a detection concentration of 5.9 10-6 ng/μl DNA for W. ceti, which is 100 times more sensitive than traditional PCR. Therefore, LAMP with PR is applicable for disease detection at the primary stages of the infection or when the clinical symptom is not obvious. The related studies have also provided effective evidence for the high sensitivity of LAMP with PR.39,40 Furthermore, the sensitivity of LAMP did not appear to be influenced by other impurities. This method also exhibits greater tolerance to interfering substances such as blood, serum, and food ingredients compared to traditional PCR.41,42 LAMP detection can even be accomplished without the DNA extraction step.43
The advantages mentioned above make LAMP more reliable for field applications. The minimum concentration of detection for W. ceti in infected rainbow trout by LAMP was assessed using boiled homogenates from kidney, spleen, and liver tissues. The findings indicated that LAMP could monitor a minimum concentration ranging from 2.1 CFU/reaction to 18 CFU/reaction in clinical applications. In a previous assay, LAMP proved a minimum concentration of 9 102 CFUmL-1 Staphylococcus aureus in milk.31 For Aeromonas salmonicida in turbot, the minimum concentration of LAMP was less than 30 CFUmL-1 in liver, spleen and kidney tissues.44 These thresholds of detection proved that LAMP was suitable for POCT or field diagnosis.
Conclusions
LAMP is a rapid and convenient molecular tool characterized by high specificity and sensitivity for the detection of W. ceti in rainbow trout. The whole procedure can be accomplished within 30 min, at 63 °C. Using boiled homogenates from liver, spleen, and kidney tissues as templates, LAMP demonstrated a minimum detection concentration of 18, 2.1, and 4.4 CFU/reaction, respectively. Therefore, this diagnostic method is a suitable tool at the early stage of infection and can provide an effective reference for the prevention and control of W. ceti in farms, especially in remote and underdeveloped regions.
Acknowledgments
This research was financially supported by the “First Class Fishery Discipline” programme[(2020)3] in Shandong Province, Natural Science Foundation of China (31902408), and Natural Science Foundation of Shandong Province (ZR2023MC154).
Authors’ Contribution
Methodology: Jiankun Pan, Yingfei Wang, Ye Tao; Formal analysis and investigation: Minghao Wang, Dandan Qian, Chaoli Zheng; Writing - original draft preparation: Huahua Fang; Writing - review and editing: Yanling Sun.
Competing of Interest – COPE
All of the authors have no conflict of interest.
Ethical Conduct Approval – IACUC
This study was accepted by the Institutional Animal Care and Use Committee of Qingdao Agricultural University, and it did not contain any studies with human participants.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
There was no data generated or analyzed in the present study.


_electrophoretic_results_of_the_.png)
_electrophoretic_results_of_the_amplification_produc.png)
_and_pcr_(b)_reactions_were_accomplished_at_different_concentra.png)
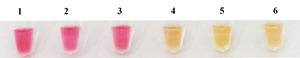


_electrophoretic_results_of_the_.png)
_electrophoretic_results_of_the_amplification_produc.png)
_and_pcr_(b)_reactions_were_accomplished_at_different_concentra.png)
