Introduction
The clam Cyclina sinensis is an important economic shell mollusk in the coastal areas of East Asia.1 Its fast growth rate and strong resistance significantly contribute to the Chinese shellfish industry.2 In recent years, ammonia nitrogen accumulation has seriously threatened marine animals’ growth and development, physiological metabolism, and behavioral responses.3
Ammonia nitrogen is an important stressor in seawater, consisting mainly of non-toxic NH4+ and more toxic NH3.4 NH3 can cause blood ammonia toxicity, which further destroys its immune system, disturbs its osmotic pressure balance, and seriously affects its growth and development performance.5 It has been shown that some aquatic animals can regulate the metabolic pathway of detoxification and change excretory behaviors to reduce toxicity.6 Current studies have shown that glutamine synthesis is one of aquatic animals’ most important detoxification pathways.7 Glutamine is a non-toxic compound that can be accumulated in large quantities in organisms.8 It can also be used to synthesize a variety of substances, such as purine, pyrimidines, and polysaccharides, which can be used in the physiological activities of the organism.9
Glutamate is synthesized by Glutamate dehydrogenase (GDH) catalyzing the synthesis of NH4+ and α-Ketoglutaric acid (α-KG), and the α-KG produced in this process can enter directly into the tricarboxylic acid cycle to provide energy for the organism.10 And glutamate can be catalyzed with NH4+ by glutamine synthetase to form glutamine.11 The reaction is considered one of the most important mechanisms of ammonia assimilation.12,13 Thus, the activity of GDH influences the synthesis of Glutamine. Recently, the function of GDH to deal with stress factors has become a research hot-spot, such as temperature,14 salinity,15 hypoxia,16 and so on. Synthesis of glutamine to reduce ammonia nitrogen toxicity has been found in Oncorhynchus mykiss,17 Monopterus albus,18 Penaeus monodon,19 Clarias gariepinus6 and so on. The function of the GDH gene in response to ammonia nitrogen has not been studied in clam. Therefore, to elucidate the functions of the GDH gene in the clam exposed to ammonia, the full-length cDNA was cloned and characterized, and ammonia nitrogen tolerance traits in the CsGDH were analyzed.
Materials and Methods
Animal maintenance and ammonia nitrogen stress
The clam C. sinensis was provided by Lianyungang Zhongchuang Aquaculture Co. The clam with basically the same specification, health, and weight (7.08±0.61g) were selected for this experiment. Before the experiment, all the clam were kept in PVC tanks (42cm×28cm×36cm) for one week to acclimate to the laboratory conditions (temperature: 21±0.5℃; salinity: 25±0.5 practical salinity units, PSU; pH: 7.77; DO>5.0 mg/L; TAN<0.01 mg/L). The water was continuously aerated, with 50% seawater replaced every 24 hours. All specimens were fed Chlorella vulgaris. The clam were fasted for 48 hours before the start of the experiment to ensure that intestinal contents had been expelled.13
The ammonia nitrogen level was set according to the previous research in the laboratory.20 Three groups were set up for the experiment, the control group (TAN: <0.01 mg/L) and the ammonia nitrogen stress group (TAN: 200 mg/L) (one group was used for the hepatopancreas tissue expression experiments, and one group was used for the correlation analysis experiments of the ammonia nitrogen tolerance traits). Experimentally added total ammonia nitrogen was configured from amorphous ammonium chloride, ready to use. Transiently reared healthy C. sinensis were randomly transferred into each experimental group, three parallels per group, and each parallel group was randomly assigned 100 C. sinensis. The water was changed once a day during the period of stress in more than 2/3 of the water volume and reconfigured after the water change.
Sample collection
To determine the expression of CsGDH in different tissues, mantle, hepatopancreas, gill, foot, pipe, and adductor were selected and removed carefully. All the selected samples were immediately frozen in liquid nitrogen. To analyze the CsGDH transcript level in the hepatopancreas of the clam exposed to ammonia nitrogen, three clams were randomly taken from each group at 0, 3, 6, 12, 24, 48, and 96 h. The hepatopancreas were removed carefully and frozen immediately in liquid nitrogen for RNA extraction.
For correlation analysis of ammonia nitrogen tolerance traits, all dead individuals were collected at 3 h intervals on patrol and numbered to indicate the time of death. Ammonia nitrogen stress was applied until 70% of the individuals died; the first 70% were ammonia nitrogen-sensitive individuals, and the last 30% were nitrogen-tolerant individuals. The hepatopancreas were removed carefully and frozen in liquid nitrogen immediately.
Cloning and sequencing of CsGDH
Gene-specific primers (Table 1) based on the putative unigenes were designed using Primer Premier 5.0 to obtain the full-length cDNA of CsGDH.13 First-strand cDNA for RACE was prepared using the HiScript-TS 5′/3′ RACE Kit (Vazyme, China).21 The 5′-RACE and 3′-RACE PCRs were performed following the manufacturer’s instructions, all operations are performed on ice.
Bioinformatic analyses of CsGDH
The full-length cDNA sequence of CsGDH was spliced and translated into amino acids using DNAMAN. Physicochemical properties of the predicted CsGDH protein were determined using the Expasy Protein Analysis System. The domains, secondary, and three-dimensional (3D) structure of the CsGDH protein were modeled with the SMART, NSPA and Swiss-Model Workspace, respectively.13 The amino acid homology analysis was performed using Esprit and CLUSTALW. A neighbor-joining (NJ) phylogenetic tree was constructed using Molecular Evolutionary Genetics Analysis (MEGA) v6.0.22
Characterization of the tissue distribution of CsGDH
To analyze the transcript level of CsGDH mRNA, qRT-PCR was performed.4 The primers used are listed in Table 1. Each sample was in triplicate. Relative transcript levels were determined using the 2-△△Ct method.4
Expression analysis of CsGDH under ammonia nitrogen stress
The expression of CsGDH gene in the hepatopancreas was detected in the clam exposed to ammonia nitrogen at different time. The reaction system, reaction procedure, data processing, and analysis were as described above.
Screening and analysis of SNPs associated with ammonia nitrogen tolerance traits in the CsGDH
The cDNA templates of 10 ammonia nitrogen-stressed individuals and ten control individuals were mixed separately. SNP primers were designed, and a single band with high specificity was selected and sent to Beijing Tsingke Biotech Co (Tsingke Biotech, China) for sequencing. The loci with double peaks in the sequencing peak plots might be SNP (Fig. 1). The sites with different bases in the sequences of the ammonia nitrogen stress group and the control group may also be SNP sites.23 The significance of the detected SNP was analyzed using SPSS 22 chi-square test. The association of SNPs with ammonia nitrogen tolerance was determined.
Results
Analysis of the CsGDH cDNA sequence
As shown in Fig. 2, the full-length cDNA of CsGDH (GenBank accession OR607255.1) was 1683 bp, including a 5′-terminal untranslated region (5′-UTR) of 459 bp and a 3′-UTR of 48 bp. The ORF of CsGDH was 1176 bp in length, encoding a protein of 391 amino acids, with a predicted molecular weight of 42.95 kDa and a theoretical pI of 6.57. Glycine accounted for the largest proportion at 10.7 percent, followed by isoleucine and alanine at 8.7 percent and 8.4 percent, respectively. The chemical formula of this protein is C1922H3018N518O570S14, and the protein instability coefficient is 24.97. As shown (Fig. 3A), the CsGDH gene includes an ELFV_dehydrog (IPR006096) functional domain located at 98 aa-387aa. Prediction of the secondary structure of CsGDH showed that the protein has 41.94% α-helices, 7.16% extended strands, and 50.90% irregular coils (Fig. 3B). The 3D structure prediction of CsGDH protein is shown in Fig. 3C.
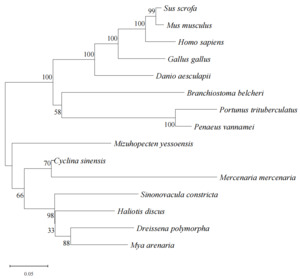
Amino acid sequence comparison, homology analysis, and phylogenetic analysis of CsGDH
The amino acid sequences deduced from C. sinensis CsGDH were compared in NCBI with the amino acid sequences encoded by GDH of other species in the GenBank database for affinity. The vertebrates such as Homo sapiens and Branchiostoma belcheri; the arthropods such as Penaeus vannamei and P. trituberculatus; Sinonovacula constricta, Mercenaria mercenaria and some other molluscs were selected for systematic analysis. A phylogenetic tree was constructed and amino acid homology comparison was carried out by using the NJ method (Fig. 4). The evolutionary tree was mainly divided into two main branches, in which C. sinensis and M. mercenaria were in a cluster, indicating that the phylogeny of these two species was the closest, and the homology with Gallus Gallus, Sus scrofa, human, and mouse species was the lowest.
Expression patterns of the CsGDH mRNA in different tissues of C. sinensis
The qRT-PCR showed that the CsGDH mRNA expressed ubiquitously in all the selected tissues. The CsGDH mRNA transcript level was highest in the hepatopancreas, followed by the gill, adductor, foot, pipe, and mantle. The CsGDH mRNA transcript level was significantly higher than other tissues (P<0.05) (Fig. 5).
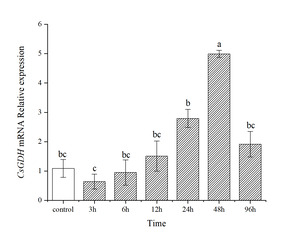
Changes in the CsGDH transcript level in the hepatopancreas of C. sinensis exposed to ammonia nitrogen
Ammonia nitrogen stress had a significant effect (P<0.05) on the transcript level of the CsGDH mRNA in hepatopancreatic tissues of the clam C. sinensis (Fig.6). The transcript level of the CsGDH mRNA in hepatopancreatic tissues of the experimental group in the first 24 h was not significant compared with that of the control group (P>0.05). The transcript level of the CsGDH mRNA was down-regulated at 3 h. And then, the transcript level of the CsGDH mRNA showed an increasing trend and reached the maximum at 48 h, which is significantly higher than the other sampling time (P<0.05).
Analysis of SNP distribution of CsGDH and its association with ammonia nitrogen tolerance
At the beginning of the ammonia nitrogen stress experiment, the first dead C. sinensis was found at 6 h. The mortality rate gradually increased with time, reaching the highest value of mortality at 96 h. At 120 h, the mortality reached 70%, which was regarded as the sensitive group, and the surviving individuals were regarded as the tolerant group. In addition, no death was found in the control group during the whole experiment. A total of seven SNPs (264bp A/G, 435bp A/G, 504bp T/A, 582bp C/T, 648bp C/T, 933bp C/T, 969bp A/G) were found, all located in the ORF region. The association analysis of ammonia nitrogen tolerance traits was performed, and the SNPs were significantly associated with ammonia nitrogen tolerance when P<0.05 (Table 2). The 504bp T/A and 582bp C/T SNPs, which were synonymous mutations, were significantly associated with ammonia nitrogen tolerance.
Discussion
GDH is one of the crucial enzymes in the synthesis of glutamine.24 Ammonia nitrogen is a serious toxic pollutant in aquatic ecosystems, and the synthesis of glutamine is an effective physiological response for organisms to adapt to high ammonia nitrogen.25 When ammonia nitrogen accumulates in the body, the GDH can reduce the ammonia nitrogen by converting the excess ammonia to glutamate.11 In the present study, the CsGDH mRNA expressed ubiquitously in all the selected tissues. The CsGDH mRNA transcript level was highest in the hepatopancreas, followed by the gill, adductor, foot, pipe, and mantle. In S. constricta, the GDH gene is expressed in the gill, foot, adductor, hepatopancreas, pipe, and mantle.26 The expression of the GDH gene was also relatively high in the hepatopancreas and gill of Fenneropenaeus chinensis (He et al., 2016). This is probably because the hepatopancreas tissue is the detoxification organ.27 Under ammonia nitrogen stress, the hepatopancreas would show edema, severe vacuolization, and local necrosis, which in turn can affect its detoxification function and even lead to fish death.28 Similarly, in this study, the CsGDH mRNA transcript levels in hepatopancreas changed with time under ammonia nitrogen stress, and the CsGDH gene was suppressed at the beginning of the stress, then showed a gradual increase and reached the extreme value at 48 h. The relative expression was 4.58-fold that of the control group, showing significant down-regulation. This phenomenon might be caused by the high ammonia nitrogen stress, which could also be seen from the expression of the CsGDH gene at 3 h. The transcriptional levels of the CsGDH gene in the stress group was significantly lower than that in the control group. Previous laboratory research has found that the structure of hepatopancreatic tissues changed significantly under ammonia nitrogen stress.4 With the prolongation of ammonia stress time, the expression of GDH gene increased gradually, accelerating the conversion of ammonia nitrogen and the synthesis of glutamate in the body.10 However, the free amino acid content in the hepatopancreas decreased under ammonia stress, among which the glutamate content decreased by 27%.29 This could be attributed to the amino acid catabolic capacity. In the final stages of ammonia stress, the transcript level of the CsGDH mRNA showed an increasing trend and reached the maximum at 48 h, which is significantly higher than the other sampling time (P<0.05). This indicates that ammonia stress can activate the CsGDH gene, contributing to ammonia nitrogen detoxification in C. sinensis.
SNP markers play important roles in biological processes, such as genetic linkage mapping, association analysis, population evolution, kinship identification, and diagnosis of diseases.30,31 As the third generation of molecular genetic markers, many studies have been conducted in Bivalvia to show that SNPs are associated with economic traits, including growth, disease resistance, and stress tolerance.32–34 Therefore, SNP markers have been widely used as a molecular-assisted breeding tool for economic traits due to the advantages of high genetic stability and detection accuracy.35 In the Patinopecten yessoensis, a synonymous mutant SNP (c.852A>G) in the IGF2BP gene was significantly associated with growth shape.36 In Tegillarca granosa, two SNPs in Hb gene were significantly associated with drug resistance in Vibrio parahaemolyticus.37 In Hyriopsis cumingii, two SNPs, +2248T/C and +2365T/C, in the IRF-2 gene were significantly associated with resistance to Aeromonas hydrophila.38 Seven SNPs were identified in the CsGDH gene and all SNPs were located in the ORF region in the present study. Association analysis showed that the 504bp T/A and 582bp C/T SNPs were significantly associated with ammonia nitrogen tolerance. Sun26 screened SNPs in S. constricta for association analysis of ammonia nitrogen tolerance traits, and the results showed that both SNPs, c.323T>C and c.620C>T, exhibited significant association (P<0.05) with ammonia nitrogen tolerance traits. This is consistent with the results of this experiment. However, proving the exact relationship between these SNPs and ammonia nitrogen tolerance traits still requires further research. On the other side, the selection and breeding of new ammonia nitrogen-tolerant C. sinensis varieties is still in its infancy, so the results of the present experiment suggest that the CsGDH gene can be used as one of the candidate marker genes to improve ammonia nitrogen tolerant of the clam C. sinensis. These data enriched the C. sinensis genome database, whereas both the small sampling population and the phenomenon of population stratification caused bias, so subsequent large population calibration is needed.
Acknowledgments
This research was supported by the earmarked fund for CARS (CARS-49), Lianyungang Key Research and Development (CG2304), Jiangsu graduate Research and Practice Innovation program (KYCX2022-65; KYCX2022-23), Jiangsu Marine Resources Development Technology Innovation Center open fund (LWJJ-01), and the Priority Academic Program Development of Jiangsu. The funding bodies played a key role in the study’s design, data collection, analysis, and interpretation, and manuscript writing.
Authors’ Contribution
Conceptualization: Hongxing Ge (Equal), Zhiguo Dong (Equal). Methodology: Hongxing Ge (Lead). Writing – review & editing: Hongxing Ge (Lead). Visualization: Jidong Hu (Lead). Investigation: Jidong Hu (Equal), Zhen LI (Equal). Formal Analysis: Qian Ni (Lead). Writing – original draft: Qian Ni (Lead). Software: Chenyu Xia (Lead). Project administration: Zhiguo Dong (Lead). Funding acquisition: Zhiguo Dong (Lead).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All are available upon reasonable request.

_mutation._b__the_582bp_(tac)_mutation.png)



_mutation._b__the_582bp_(tac)_mutation.png)




