Introduction
Cyclophilins (Cyps) are a multi-functional protein family with extensive natural distribution and highly conserved properties. They are intracellular receptors of the immunosuppressant Cyclophilin A(CsA), which is known as CSA-binding protein.1 CypA research focuses on its distribution in different organisms, analysis of related protein sequences, structure, and critical biological functions.2 Many studies have reported that CypA is closely related to the occurrence of tumors. The expression of CypA was inhibited in non-small cell lung cancer cells by RNA interference technology, and the down-regulation of CypA expression resulted in a significant decrease in the proliferation of non-small cell lung cancer cells.2 Through proteomics and cytobiology, it was found that expression levels of CypA were also significantly up-regulated in esophageal squamous cell carcinoma, male breast cancer, prostatic cancer, head-neck tumor, tongue squamous cell carcinoma, and other tumors, and CypA participated in malignant biological behaviors.3–7 The molecular chaperone function of CypA played a vital role in the repair of misfolded and broken conditional complexes, which were caused by improper signal activation in tumor formation, and it could maintain the steady state of proteins, which might be essential to the survival of tumors.8 In addition, CypA participates in virus replication as an immunosuppressor and is involved in the body’s immune response.9 Qiu et al. found that CypA played an important role in resistance to viral infection in mammals, indicating that CypA participated in the innate immune response.10 In recent years, it has been found that CypA may play an immune stress role in aquatic organisms, such as Chlamys farreri, Hyriopsis schlegelii, Eriocheir sinensis, Penaeus monodon, Litopenaeus Vannamei, Pelteobagrus fulvidraco and Ictalurus punctatus.11–15
Megalobrama amblycephala, with delicious taste, high meat content, and a small head, is deeply loved by consumers. Since promoting culture in 1960, it has become the sixth large-scale freshwater cultured fish in China.16 In recent years, with the rapid development of M. amblycephala breeding industry, pathogenic factors such as bacteria, viruses and parasites have caused rampant breeding diseases, and high-density breeding has led to eutrophication of water bodies, which has seriously threatened M. amblycephala breeding industry. Therefore, studying the immune system and its regulatory mechanism is vital for the sustainable development of M. amblycephala culture.
A. hydrophila infection is one of the major causes of large-scale death in cultivation of M. amblycephala. Due to the short onset time and high mortality rate, A. hydrophila infection has become a constraint in the green development of the M. amblycephala aquaculture industry. Nevertheless, the immune response mechanism of CypA in different tissues of M. amblycephala after A. hydrophila infection remains unknown. Therefore, this study aimed to clone the full-length cDNA of M. amblycephala CypA for sequence analysis, and compare it with the homologous sequences of other species to construct the phylogenetic evolutionary tree. Expression levels of CypA in different tissues of M. amblycephala were detected by quantitative real-time PCR (qPCR). After A. hydrophila infection, expression levels of CypA were detected in various tissues of M. amblycephala. The results of this study will provide basic experimental data to study disease control and anti-infection mechanisms of M. amblycephala.
Materials and Methods
Experimental M. amblycephala
M. amblycephala (50±2 g) were collected from the aquatic breeding base at the Hunan Applied and Technology University. M. amblycephala were cultured in the laboratory cyclic aquaculture system for 14 d before sampling, during which aeration with oxygen was supplied for 24 h per day to keep enough oxygen dissolved in the water. The water temperature was 26±2 °C. The 100 mg tissues of liver, spleen, head kidney, intestines, gills, kidneys, and heart were collected from healthy M. amblycephala, respectively. All tissues were frozen in liquid nitrogen and stored in a refrigerator at a temperature of -80 °C.
A. hydrophila infection
A. hydrophila was acquired from the Hunan Applied and Technology University microbiology laboratory. Monoclonal strains were selected, inoculated in the Luria-Broth (LB) liquid medium, and cultured for 18 h by shaking at 28 °C. The solution was then centrifuged at a rate of 4000 rpm for 10 min, and the supernatant was removed. Next, the remaining sample was diluted via sterile PBS solution, and the concentration of bacteria after dilution was determined by plate count. Meanwhile, the concentration of bacteria was diluted to 1.0×109 CFU/mL for later use. Three parallel repeating groups were established with 30 M. amblycephala samples in each group. All M. amblycephala samples were immersed in aquaculture boxes with 1.0×109 CFU/mL A. hydrophila for 30 min, and then transferred into breeding tanks with aeration. Samples were collected at 3 h, 6 h, 12 h, 24 h, 48 h, and 72 h, respectively. Healthy M. amblycephala samples without infection were chosen as the control group. In each sampling process, 5 M. amblycephala samples were collected from the experimental and control groups. 100 mg each of gill, intestine, liver, head kidney, and spleen tissues were collected, respectively.
Total RNA extraction and reverse transcription
Utilizing the Simgen animal tissue total RNA extraction kit, total RNA of M. amblycephala liver was extracted. Concentration and purity of RNA (OD260/OD280) were tested using the nucleic acid concentration tester. The integrity of RNA was tested by electrophoresis and an RNA sample with clear bands and good integrity was chosen as the template. Reverse transcription was carried out using the Simgen reverse transcription kit. The acquired cDNA of M. amblycephala liver was stored at a temperature of -20 ℃ for later use.
Amplification of full-length cDNA sequences of M. amblycephala CypA
According to the cDNA sequences of Cyprinidae fish CypA found in the NCBI database, primers (Table 1) were designed using Premier 5. Amplification of the middle sequences of CypA used cDNA extracted from liver tissues as the template. The PCR reaction conditions were as follows: 94 °C, 5 min; 94 °C, 30 s; 54 °C, 30 s; 72 °C, 1 min; 30 cycles; 72 °C, 10 min; and finally, terminated at 4 ℃. PCR products were recycled and purified. After linking, transformation, and PCR identification, the positive bacterial liquid were sequenced.
According to the middle sequence of the CypA of M. amblycephala gained from sequencing, 5′ and 3′ RACE primers (Table 1) were designed. Using the total RNA of the liver as a template and RACE kit (Takara, Japan), the two ends of the CypA of M. amblycephala were amplified. The PCR conditions were as follows: 95 ℃, 3 min; 95 ℃, 30 s; 72 ℃, 2 min; 8 cycles; 95 ℃, 30 s; 68 ℃, 30 s; 72 ℃, 2 min; 6 cycles; 95 ℃, 30 s; 64 ℃, 30 s; 72 ℃, 2 min; 30 cycles; extension at 72 ℃ for 5 min; and finally, terminated at 4℃. After recycling and purification, PCR products were connected to a linear pRACE vector and then transformed into the competent E. coli DH5α cells. The positive bacterial liquid was sequenced after screening by culture medium, followed by PCR identification. The acquired cDNA segments were spliced for sequence analysis and comparison. The ORF primers were designed further to verify the cDNA sequences of M. amblycephala CypA.
Sequence analysis
Using DNAstar software, the cpmpleted cDNA sequence of CypA of M. amblycephala was acquired through splicing. In NCBI database (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), utilizing the online ORF finder, the amino acid sequence of CypA of M. amblycephala was predicted. In GenBank, using the Blastn tool, the homologous sequences of CypA were searched. The amino acid structural domains of CypA were annotated using UniProt and SMART. Using the SWISS-MODEL, the tertiary structure of M. amblycephala CypA was predicted. Multi-sequence comparison was analysed with Clustal X 1.83, and the phylogenetic tree of the genetic system was established with MEGA 5.1.
Relative expression of M. amblycephala CypA
Different tissues of M. amblycephala were collected. RNA was extracted using the Simgen animal tissue total RNA extraction kit. Utilizing the degenome reverse transcription kit (TaKaRa), the cDNA template was obtained by reverse transcription. According to M. amblycephala CypA sequenc, qPCR primers (Table 1) were designed, and the reference gene was β-actin gene. Expressions of CypA in different tissues of M. amblycephala were detected using a qPCR kit (TaKaRa). The qPCR amplification conditions were as follows: 95 ℃, 5 min; 95 ℃, 20 s; 56 ℃, 20 s; 72 ℃, 20 s; 40 cycles; 72 ℃, 8 min. The relative expression level of M. amblycephala CypA in different tissues and at various time after immersion infection were calculated via the ΔΔCT method.
Data analysis
Experimental results are expressed as mean ± standard error. Different expression levels of CypA in different tissues of M. amblycephala and at different time points after infection were examined through one-way analysis of variance (ANOVA) using SPSS statistical software. The significance level is P< 0.05.
Results
Full-length cDNA sequences of M. amblycephala CypA
The complete cDNA sequence of M. amblycephala CypA is 901 bp (GenBank ID: PV256473). The 5′ noncoding region is 77 bp. The 3′ noncoding region is 329 bp, which includes a termination codon and a poly-A tail. The open reading frame is 495 bp and it encodes 164 amino acids. The molecular weight of CypA is 17.43 KDa. The theoretical isoelectric point is 8.6. The molecular formula of M. amblycephala CypA amino acid sequence is C768H1202N214O230S10, which contains 16 acidic amino acids (Asp+Glu) and 19 alkaline amino acids (Arg+Lys). The extinction coefficient is 8730 and the absorption coefficient is 0.501. The instability coefficient is 0.57, indicating that CypA is a very stable protein (Figure 1).
Homology analysis of CypA in M. amblycephala
According to comparative analysis of CypA with other fish, the similarity was as follows: Cyprinus carpio (94.51%), Ctenopharyngodon idellus (92.07%), Carassius gibelio (92.07%), Labeo rohita (92.07%), Danio rerio (91.46%), and Onychostoma macrolepis (91.46%) (Table 2). Multiple sequence comparisons between the amino acids of CypA in M. amblycephala and those of seven other species was carried out. Results showed that the amino acid sequence of CypA in M. amblycephala was similar to that of other species, and all had a peptidyl prolyl cis-trans isomerase (PPIase) family tag sequence (FKGSAFHRIIPGFMCQGG). Moreover, they also contain essential amino acid sequences (Arg55, His54, Phe60, Phe113, His126) for PPlase activity and 13 conservative CSA binding residues in the exact location (Figure 2).
Tertiary structure of M. amblycephala CypA
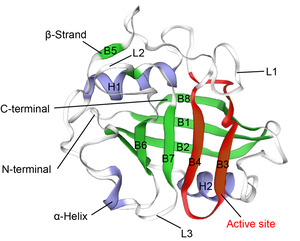
The tertiary structure (PDB id: 2z6wB) of CypA reported by Kallen et al.17 was used as a template, and the tertiary structure of CypA in M. amblycephala was gained through ESyPred3D prediction (Figure 3). The tertiary structure was analyzed according to prediction results. It was found that CypA in M. amblycephala contained 8 β-pleated sheets and 2 α-helixes, which formed a cylinder structure (Figure 3). It was speculated to form an active center region. Additionally, the tertiary structure of CypA in M. amblycephala shows that the Phe48~Gly65-bit amino acid sequence (FKGSAFHRIIPGFMCQGG) forms a divergent loop structure, which is the characteristic sequence of CypA.
Evolutionary tree of the genetic system of CypA
The predicted amino acid sequence of M. amblycephala CypA was compared with those of other species, and the phylogenetic tree was constructed by the adjacency method. M. amblycephala CypA is clustered with CypA of Cyprinidae fish, and CypA of M. amblycephala is most closely related to CypA of Cyprinus carpio (Figure 4).
Expression of CypA in different tissues of M. amblycephala
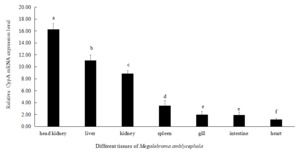
The mRNA expression level of M. amblycephala CypA was highest in the head kidney, followed by liver, kidneys, and spleen. There is no significant difference between the mRNA expression levels in the gills and intestines. The relative expression level of CypA in the heart of M. amblycephala was the lowest (Figure 5).
Immune response of M. amblycephala CypA to A. hydrophila
M. amblycephala was infected by immersion with A. hydrophila. The CypA expression first increased and then decreased in the first 72 h of infection in different tissues (Figure 6). The mRNA level of CypA in the liver increased significantly at 3 h (1.73-fold), reached its peak at 12 h (6.89-fold), and then declined gradually. By 72 h, it had decreased to 2.25-fold that of the control group. The relative expression of CypA in head kidney increased significantly at 3 h (5.79-fold), reached its peak at 6 h (15.02-fold), and then decreased gradually at 12 h. The expression of CypA in gills peaked at 12 h (7.42-fold) and then decreased gradually. The expression level in gills decreased to 1.52-fold that of the control group at 72 h. The relative expression of CypA in intestines reached its maximum at 24 h, which was 9.92-fold that of the control group, and then decreased to 3.69-fold that of the control group at 72 h (Figure 6).
Discussion
Cyclophilins (Cyps) are a type of multi-functional protein family with extensive natural distribution and highly conserved properties. This study obtained CypA from M. amblycephala through cloning for the first time. The completed cDNA sequence of M. amblycephala CypA was 901 bp with a 495 bp-long open reading frame at 78-572 bits, which encodes 164 amino acids. The sequence also includes a 77 bp 5′ noncoding region, a 329 bp 3′ noncoding region, and a 30 bp poly(A) tail. However, no tailing signal has been found yet. Through multi-sequence comparative analysis, M. amblycephala CypA contained a PPIase active site. In both vertebrates and invertebrates, this site is highly conserved, and is located at the 48th to 65th amino acids (FKGSAFHRIIPGFMCQGG). The conservative amino acid residues in M. amblycephala CypA are crucial to the level-1 structure of CypA sequences and PPlase activity, which are related to other biological functions of CypA.18,19 CypA of M. amblycephala is a kind of PPIase and it binds with immunosuppressor CSA as an intracellular receptor.
According to the prediction of tertiary structure, it was found that M. amblycephala CypA is composed of 8 β-pleated sheets and 2 α-helixes. Moreover, the CypA structure of M. amblycephala showed that the amino acid sequence (FKGSAFHRIIPGFMCQGG) at the 48-65th amino acids formed a divergent loop structure and thereby developed a characteristic sequence of cyclophilin. The evolutionary tree of the genetic system demonstrated that CypA in Megalobrama amblycephala is clustered with CypA in Cyprinus carpio, indicating the closest parentage between them.
Studies on CypA of vertebrates demonstrated that these proteins participate in many biological processes. CypA is involved in immune defense in mammals after viral infection and play a key role in pathogenesis of some inflammatory diseases in humans.20 Nevertheless, in innate immune systems of aquatic animal, there exists little knowledge concerning the fuction of CypA. Xu Ling detected expression distributions of CypA in various tissues by qPCR and found that expressions of CypA in the liver and gonads of Hyriopsis Schlegelii were significantly higher compared to those in other tissues.21 Moreover, mRNA expression levels of H. Schlegelii CypA in heart and kidney are relatively higher than those in other tissues.21 The mRNA level of H. Schlegelii CypA was lowest in intestines, and therer had no significant difference in the gills, axe foot, intestines. and hemolymph.21 CypA is expressed in the hepatopancreas, ovaries, muscles, heart, brain, and hemolymph of Penaeus monodon.10 Similar results have been observed in Chlamys farreri; the mRNA level of Chlamys farreri CypA is highest in gonads, but its expression in muscles and hemolymph is the lowest.9 In M. amblycephala, CypA is expressed in the head kidney, liver, intestines, gills, spleen, kidneys, and heart, and its expression in head kidney is the highest, followed by liver, kidneys, spleen, gill and intestines. The relative expression of CypA in the heart of M. amblycephala is the lowest. It is suggested that M. amblycephala CypA might play a key role in the head kidney.
Infected by A. hydrophila, no significant changes were found in terms of mRNA expression of CypA in the hemolymph of Hyriopsis Schlegelii.21 However, mRNA expression level of H. Schlegelii CypA in the gonads increased significantly at 4 h after infection (P<0.05) and then reached a peak.21 Expression of CypA in the gonads of H. Schlegelii begins to increase 2 h after infection. Still, it decreases after 8h and reaches a valley at 24 h, after which it increases partially at 48h.21 A similar law has been found in the liver of H. Schlegelii, after infection by A. hydrophila, the expression of CypA increased significantly at 4h and then reached a peak.21 The mRNA expression level of H. Schlegelii CypA in the liver increased at 2h after infection, then decreased at 8 h and reached a valley at 48h.21 This indicates that expression of CypA might be induced by A. hydrophila in the gonads and liver of H. Schlegelii, which was in line with expression of CypA in the gonads of Chlamys farreri after infection by Vibrio anguillarum.9,21 After infection by Edwardsiella tarda, the expression level of Ictalurus punctatus CypA in egg cell lines was upregulated, which indicated that CypA might play a key immune role in the early development stage of Ictalurus punctatus infected by Edwardsiella tarda.15 Aafter LPS stimuli, expression of Penaeus monodon CypA in the hepatopancreas was upregulated.10 To further explore the potential functions of CypA in M. amblycephala, M. amblycephala was immersed in A. hydrophila for infection. The expression level of CypA in the head kidney, liver, gills, and intestines first increased and then decreased in the first 72 h after infection. Moreover, the expression level of Megalobrama amblycephala CypA in the head kidney peaked at 6 h. The mRNA level of CypA peaked at 12h in gills and liver and 24h in intestines. This demonstrates that A. hydrophila can stimulate CypA expression in M. amblycephala, and CypA might participate in the immune defense of fish against exogenous microbiological stimuli.
After infection by A. hydrophila, the mRNA level of CypA in the head kidney of M. amblycephala reached the peak (6 h) earlier than in the liver (12 h) and intestines (24 h). Moreover, the relative expression level of Megalobrama amblycephala CypA mRNA at peak was also higher in the head kidney than in liver, intestines, and gills. It was suggested that CypA might play a key role in resisting bacterial infection in the head kidney. Nevertheless, the special effect of CypA in the head kidney is still unknown, and further studies are needed to obtain a more comprehensive understanding. Further learning on CypA is conducive to exploring its functions in fish and understanding the mechanism of fish immune responses. This is also beneficial for a better understanding of the immune response mechanism of fish.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 32360922), the Guangxi Natural Science Foundation (Grant No. 2022JJA130338), the Hunan Natural Science Foundation (Grant No. 2025JJ50145), and the first-class specialty in Aquaculture of Hunan Applied and Technology University (HYYLZY202002).
Authors’ Contribution
Conceptualization: Juanjuan Lu (Equal), Hu Xia (Equal). Methodology: Juanjuan Lu (Equal), Hu Xia (Equal), Yanan Gong (Equal). Writing – original draft: Juanjuan Lu (Equal), Hu Xia (Equal). Formal Analysis: Hu Xia (Equal), Fuyan Chen (Equal), Jianchao Bu (Equal), Pinhong Yang (Equal). Investigation: Hu Xia (Equal), Fuyan Chen (Equal), Jianchao Bu (Equal), Pinhong Yang (Equal). Writing – review & editing: Hu Xia (Equal). Funding acquisition: Hu Xia (Lead). Resources: Hu Xia (Lead). Supervision: Hu Xia (Lead).
Competing Interests – COPE
The authors of this study declare no conflict of interest.
Ethical Conduct Approval – IACUC
The experiment was conducted with approval from the IACUC (Institutional Animal Care and Use Committee) of Hunan Applied and Technology University, Changde, China.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All are available upon reasonable request.