Introduction
Over the past decades, conservation genetics has emerged as a pivotal tool for managing and recovering endangered taxa.1 In genetic studies of imperiled species such as Gymnocypris chilianensis (Cyprinidae), the reliability of DNA sampling methods and their impact on animal welfare are critical considerations. Traditional destructive sampling, which requires euthanizing animals to extract tissues, is unsuitable for endangered species.2 Consequently, non-invasive sampling has gained prominence. This approach collects genetic material from naturally shed biological samples (e.g., mucus, feces, scales, or skin/oral swabs) without capturing or disturbing individuals.3,4 Non-invasive sampling has been successfully applied to endangered species like the Chinese Egret (Egretta eulophotes),5 Ocean Sunfish (Mola mola),6 and Chinese Giant Salamander (Andrias davidianus).7 However, terminological inconsistencies persist: some studies broadly classify minimally invasive techniques (e.g., skin swabbing) as “non-invasive” (sensu lato), while stricter definitions (sensu stricto) mandate no physical contact.4 In contrast, non-lethal methods (e.g., fin clipping) retain animal survival but may impair behavior and fitness due to invasiveness.4,8 Long-term comparative analyses of sampling impacts—such as growth metrics (length-weight relationships, condition factor) and mortality—are thus essential to optimize ethical and practical outcomes.9,10
Gymnocypris chilianensis (Cyprinidae) was first named by Li and Zhang (1974)11 during a fish survey in the Hexi Corridor area. The distribution of G. chilianensis is very narrow in China, it distributed only in the three rivers originating from Qilian Mountains, including Shiyang River, Heihe River, and Shule River.12 In recent years, due to changes in the ecological environment, construction of water conservancy projects and human overfishing, the habitat of G. chilianensis has been severely damaged, resulting in a sharp decrease in wild resources.13 Therefore, the protection of its wild germplasm resources is particularly important. Research on genetic diversity based on DNA serves as a crucial prerequisite for formulating effective conservation strategies for G. chilianensis, and thus has garnered widespread attention.14 Currently, research on G. chilianensis primarily involves collecting small amounts of muscle tissue from live specimens, with limited studies utilizing non-invasive sampling methods.
The evolution speed of Cytb gene is moderate, and it is widely used to study the genetic diversity and genetic structure of populations.15 The mitochondrial D-loop region, lacking coding constraints, is the most rapidly evolving region in mitochondrial DNA due to the absence of selective pressure. It is often used to study the genetic differentiation between populations and within populations.16
Here, we evaluate long-term effects (70 days) of skin swabbing versus fin clipping on growth performance and mortality in farmed G. chilianensis. Concurrently, mitochondrial Cytb and D-loop sequencing will elucidate genetic diversity patterns to inform breeding strategies and mitigate inbreeding risks.17,18 This dual approach aims to provide reference for the breeding and germplasm improvement of good varieties of G. chilianensis, to advance ethical sampling protocols and genetic conservation frameworks for G. chilianensis, and will be beneficial to the conservation and sustainable utilization of threatened freshwater fishes.
Materials and Methods
Experimental materials and sources
The G. chilianensis individuals used in the experiment were all artificially bred in a farm named Gansu Cold Water Fish Breeding Center (Linze, Gansu, China), which is a typical plateau area with an altitude about 1500 meters high above sea-level.19
Given that seventy days generally spans the essential developmental milestones in piscine growth cycles, this temporal span was accordingly designated as the study period.
Experimental design
A total of 180 individuals of varying sizes were randomly selected and divided into three groups: “Group N” for skin swabs, this group represents non-invasive sampling. “Group Q” for partial pectoral fin samples, and “Group C” as the control group, with 60 individuals in each group. Group N (mucosal biopsy via dermal swab) represents non-invasive monitoring. Group Q (sublethal fin ray excision) represents non-lethal sampling. Group C (baseline control) received no manipulative procedure. We denote the beginning data of the experiments as “A” and the ending data of the experiments as “B”. Consequently, we have six sets of data: CA, QA, NA, CB, QB, and NB. The samples from the three groups were separated into nine tanks (20 individuals in each tank) and cultured in the same environment, including water temperature, pH value, dissolved oxygen, and feeding methods. On March 8, 2023, the standard length and weight of all individuals in each group were measured, recorded, and then released into the tank. On May 18, 2023, after 70 days of culturing since March 8, the standard length and body weight of all individuals, as well as the number of surviving individuals were measured and recorded again.
Genomic DNA Extraction
Using a sterile swab, gently wipe the surface mucus on one side of the skin three times. Then place the swab into a 1.5 mL EP tube. Add 100 μL of lysis buffer to the EP tube. The lysis buffer contains 100 mmol/L NaCl、10 mmol/L Tris-HCl、1 mmol/L EDTA (pH = 8.0)、1.0% SDS、0.1 mg/mL proteinase K. Incubated at room temperature for 15 minutes to complete lysis. Genomic DNA was extracted from each sample using an Animal Tissue Genomic DNA Extraction Kit by Sangon Biotech (Shanghai) Co., Ltd. DNA concentration was determined using a nucleic acid/protein analyzer (Merinton spectrophotometer), and DNA quality was assessed through 1% agarose gel electrophoresis.
PCR Amplification and Sequencing
Primer Premier 5 software20 was used to design primers for amplifying the Cytb gene sequence and D-loop region sequences. The primer information used in this study is detailed as follows: for the amplification of cytochrome b gene (cytb), L14724 (forward primer, 5’- GACTTGAAAACCACCGTTG -3’) and H15915 (reverse primer, 5’- CTCCGATCTCCGGATTACAAGAC -3’) were selected to form a primer pair; The mitochondrial control region (D-loop) was amplified by D-loop F (forward primer, 5 ‘- GGGATATGTCATCCTTTATGG -3’) and D-loop R (reverse primer, 5 ‘- GGGTTTGACAAGAATAACAGG -3’). The PCR reaction system (25 μL total volume) consisted of 1 μL dNTP Mix, 1 μL each of forward and reverse primers (10 mmol/L), 1 μL template DNA, 2.5 μL Taq Buffer, 0.2 μL Taq polymerase, and ddH2O added to reach a final volume of 25 μL. The PCR amplification parameters were set as follows: 95°C pre-denaturation for 5 min; 30 cycles of 94°C denaturation for 30 s, 58°C annealing for 30 s, and 72°C extension for 30 s; followed by a final extension at 72°C for 30 s. The PCR products were purified and subjected to bidirectional sequencing using Sanger method on ABI 3730XL Sequencer.
Sequence Alignment and Phylogenetic Analysis
Sequences were aligned using Clustal X software21 and manually corrected, with trimming of base pairs exhibiting unstable sequencing signals at both ends. Genetic diversity parameters of the cultivated G. chilianensis population were analyzed using DnaSP 6.0,22 including Tajima’s D neutrality test. Nucleotide mismatch distribution analysis was performed for both the Cytb gene and D-loop region sequences. Base composition was calculated with MEGA 11,23 and Neighbor-Joining (NJ) phylogenetic trees (based on the Kimura 2-parameter model24) were constructed for both sequence haplotypes using 1000 bootstrap replicates.25
Measurement of growth parameters
Weigh the body weight (W) using a digital electronic scale, accurate to 0.1g; Measure the standard length (L) with a ruler, accurate to 0.1cm. Based on the analysis of biological measurement data before and after the experiment, the standard length and body weight distribution of three groups of G. chilianensis samples were analyzed in GraphPad Prism.26
Relationship between standard length and body weight
Preliminary analysis (graphical plots) indicated the presence of non-linear relationships. Consequently, generalized additive models GAM,27 using the ‘mgcv’ package in R.28 The model uses the standard length of the sample as the response variable and body weight as the explanatory variable to explore the non-linear relationship between the standard length and body weight of fish.
W=s(L)+ ∈
Where W represents the weight of fish, L denotes the standard length of fish, s (▪) is a smooth function used to fit the potential non-linear relationship between standard length and body weight, and ∈ represents the error term, assumed to follow a normal distribution.
Condition Factor
Fulton state fullness (K) indicates the growth state of fish.29 The higher the K value, the better the growth of fish, and its calculation formula is:
K = W/ L3 × 100
Where W represents the weight of fish, L denotes the standard length of fish.
Mortality
Death is defined as the cessation of individual life activities in fish, characterized by the cessation of swimming, stiffening of the body and loss of mobility.30 In this experiment, the number of dead individuals in each of the three groups was recorded and analyzed using SPSS software.31 Firstly, the mortality percentage for each group was calculated (number of dead individuals/total number of individuals × 100%). Subsequently, a chi-square test (or Fisher’s exact test when the sample size is small) was employed to determine whether the difference in mortality rates between the two groups was statistically significant (P<0.05 indicates a significant difference).
Results
Characteristics and Variations of Cytb and D-loop Sequence Fragments
Consensus sequences of the Cytb gene (1098 bp) and D-loop region (696 bp) were obtained from 70 G. chilianensis individuals. Base composition and variation rates demonstrated that the Cytb and D-loop sequences of G. chilianensis in this study exhibited A+T contents of 56.3% (Cytb) and 63.5% (D-loop), the average A+T content in both sequences exceeded G+C, consistent with mitochondrial base composition in teleost fishes.
Population Genetic Structure
For the Cytb sequences, 6 polymorphic sites (S), 7 haplotypes (H), a haplotype diversity (Hd) of 0.354, nucleotide diversity (Pi) of 0.00040, and an average nucleotide difference (K) of 0.384 were identified. The D-loop region sequences revealed 10 polymorphic sites (S), 9 haplotypes (H), Hd of 0.787, Pi of 0.00315, and K of 1.841 (Table 1). Comparative analysis demonstrated that the D-loop region exhibited higher haplotype diversity (Hd) and nucleotide diversity (Pi) than the Cytb sequences. Additionally, the D-loop region showed greater numbers of polymorphic sites and haplotypes, along with a significantly higher average nucleotide difference (K). These results clearly indicate that genetic diversity metrics based on the D-loop region sequences are markedly superior to those derived from the Cytb sequences.
Population Genetic Differentiation
The average genetic distance for the Cytb gene in the cultivated G. chilianensis population was 0.00133, while the D-loop region exhibited a higher value of 0.00279, indicating a greater degree of genetic differentiation based on the D-loop region compared to the Cytb gene.
Neutrality test analysis yielded Tajima’s D values of -1.65080 (P > 0.05) for Cytb and -0.30479 (P > 0.05) for the D-loop region. Nucleotide mismatch distribution analysis of both sequences revealed unimodal patterns (Figures 1A, 1B), suggesting an excess of nucleotide site variations relative to neutral evolutionary expectations. These results may reflect a historical population expansion.
NJ Phylogenetic Trees of the G. chilianensis Population
The haplotype distributions for the Cytb gene and D-loop region are summarized in Table 2. Neighbor-Joining (NJ) trees constructed from both sequence haplotypes exhibited polytomies (Figures 2A, 2B). D-loop region: Hap1, Hap4, Hap5, and Hap6 clustered into a dominant branch, representing 46 individuals. Haplotypes Hap2 and Hap3 formed a subclade, representing 14 individuals. Haplotypes Hap8 and Hap9 first clustered together, then merged with Hap7, forming a branch representing 10 individuals. Cytb gene: Hap1, Hap2, Hap3, and Hap4 grouped into a major cluster, representing 65 individuals. Hap5 diverged as a singleton branch, representing 1 individual. Haplotypes Hap6 and Hap7 each formed distinct branches, collectively representing 4 individuals.
Statistical analysis of morphological indexes
Each group comprised 60 individuals selected through random sampling, and their standard length and body weight were recorded after a long-term observation of 70 days. The three sets of data before the experiment were denoted as CA, QA, and NA, while the three sets of data after the experiment were denoted as CB, QB, and NB. The distribution of standard length and body weight of the three groups of G. chilianensis is relatively concentrated. Before the experiment, the standard length of the samples in the three groups was mainly between 15 cm and 25 cm, accounting for 93.33%, 96.66% and 100% respectively, and the body weight was mainly distributed between 40 g and 100 g, accounting for 80.00%, 87.50% and 81.66% respectively. Perform an independent-samples T-test on the two experimental groups before and after the experiment respectively with the control group. As shown in the Figure 3, the results indicate that there is no significant difference between the two experimental groups and the control group before the experiment (P > 0.05), and likewise, there is no significant difference between the two experimental groups and the control group after the experiment (P > 0.05).
Regression relationship between standard length and body weight based on GAM
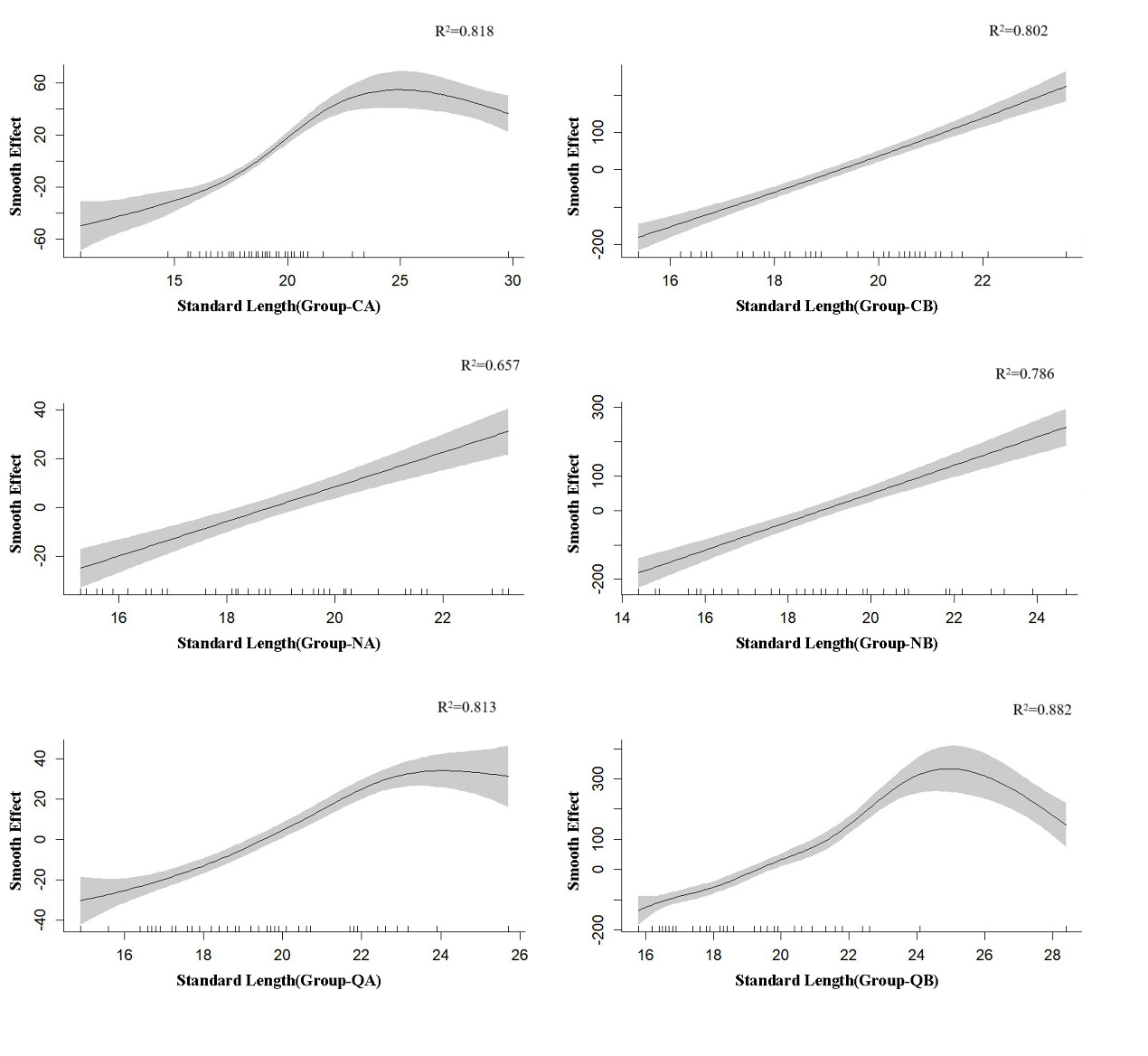
The fitting results of the regression relationship between the standard length and body weight of G. chilianensis samples before and after the experiment are shown in Figure 4. They clearly demonstrate the distinct growth patterns of the three groups of samples before and after the experiment, all exhibiting a non-linear relationship. The QB group demonstrates the best fitting effect, while the NA group shows the least. Specifically, when the standard length exceeds 25, an inflection point appears in the CA group. When the standard length surpasses 24, inflection points emerge in both QA and QB groups. However, no notable inflection points are observed in the remaining groups.
Condition Factor
The discrete distribution of sample condition factors among the three groups after the test is illustrated in the Figure 5A. The K values for all three groups exceed 200%. The lowest K value is observed in Group Q, approximately at 200%, while the highest K value is found in Group N, nearing 800%. Overall, the distribution of K values among the three groups is relatively similar, with most falling between 400% and 600%. Condition factor, as an important indicator reflecting the growth status of fish, is used to denote the growth rate of fish and the nutritional status of fish stocks. According to the fish plumpness standard established (Morton, 2006), a plumpness range of 80% to 100% is considered extremely poor, 100% to 120% is poor, 120% to 140% is fair, 140% to 160% is good, and a level of 160% or above is excellent. When applied to this study, the results indicated that the plumpness of all three groups of G. chilianensis exceeded 160% after 70 days of the experiment, indicating that the plumpness evaluation could achieve excellent levels.
Mortality
After 70 days of long-term observation, 8 fish in group C died, 9 in group N died, and 10 in group Q died. The mortality rates of the three groups were 13.33%, 15%, and 16.66% respectively (Figure 5B). Given that mortality was also observed in the control group, this likely represents natural mortality during long-term cultivation and does not affect the validity of the final experimental outcomes. After ANOVA analysis, there was no significant difference in mortality rates between the groups.
Discussion
Non-invasive sampling (sensu stricto) emphasizes sampling without physical contact with the target.4 However, since aquatic animals make it challenging to obtain DNA without such contact, this study adopted non-invasive sampling (sensu lato). Referring to latest interpretation of sampling methods, the two methods explored in this study align with the narrow definitions of both non-invasive and non-lethal sampling. This study analyzed the mitochondrial Cytb sequence and D-loop region sequence of the G. chilianensis. Based on non-invasive sampling techniques, the genetic characteristics of the aquaculture population were revealed, compared the long-term effects of non-invasive and non-lethal sampling on fish growth, selecting body surface wiping as a representative of non-invasive sampling and pectoral fin cutting as a representative of non-lethal sampling. In practice, providing reference for molecular ecology and conservation biology research of G. chilianensis and other rare and endangered fish species in the Qilian Mountains.
Sequence Characteristics of the Cultivated G. chilianensis Population
Mitochondrial Cytb gene and D-loop region sequences are widely used in fish population genetics. Consistent with the general characteristics of teleost mitochondrial genomes—“high A+T content and low G+C content” both exceeding their respective G+C values.
Genetic Diversity Analysis of the Cultivated Population
Compared to the wild G. chilianensis populations studied by Zhang et al.14 the cultivated population in this work showed reduced genetic diversity in both Cytb and D-loop regions, likely due to genetic bottleneck effects under artificial breeding conditions. Grant et al.32 proposed thresholds for genetic diversity metrics: haplotype diversity (Hd) ≥ 0.5 and nucleotide diversity (Pi) ≥ 0.005. In this study, the Cytb gene fell below both thresholds (Hd = 0.354, Pi = 0.00040), indicating low genetic diversity, which may reflect a founder effect from limited parental stock during early breeding stages. Rapid expansion of aquaculture scale post-initial breeding success likely amplified the genetic influence of a few founder individuals. In contrast, the D-loop region exceeded the Hd threshold (0.787) and exhibited higher Pi (0.00315), suggesting stable population genetic structure. Overall, the D-loop region demonstrated superior genetic diversity and structural stability compared to Cytb, aligning with its faster mutation rate in mitochondrial DNA. These findings confirm the D-loop’s suitability as an ideal marker for studying genetic differentiation in this species.
NJ Haplotype Tree Analysis of the Cultivated Population
The NJ haplotype trees revealed polytomies (Figure 2), reflecting complex internal genetic structure. High bootstrap values supported three primary branches across both genes, suggesting maternal origins from three distinct lineages. Wild G. chilianensis populations are distributed across three inland river basins in the Qilian Mountains: the Shiyang River, Hei River, and Shule River. Geographic barriers (e.g., mountainous terrain) and seasonal river fragmentation likely restricted gene flow between basins, leading to genetic divergence among broodstock selected from different maternal lineages. The dominance of specific branches (e.g., 65 individuals in the Cytb major cluster) implies potential adaptive advantages of certain genotypes under aquaculture conditions. Future studies should analyze phenotypic traits associated with these haplotypes to inform precision breeding strategies for new varieties.
The long-term impacts of non-invasive sampling on growth
This study uses the G. chilianensis as a representative species. After conducting non-invasive and non-lethal sampling on the samples, we evaluated whether there were significant impacts on the long-term (70 days) growth and development of the samples through standard length, body weight parameters, fatness, and mortality rates. The experiment proved that there were no significant differences in several indicators between the samples after both types of sampling and the samples without sampling, providing a strong basis for the further application of non-invasive and non-lethal sampling in ichthyology and even conservation biology research.33 The results of standard length and body weight parameters showed that there was no significant difference in the growth and development of the samples after sampling compared to the samples without sampling, indicating that the growth and development of the fish were not significantly affected. The lack of significant difference in fatness reflects that sampling has caused damage to the feeding behavior and energy reserve process of the samples. The lack of significant difference in mortality rates further demonstrates that both sampling methods pose less threat to the fish, and also proves that there was no significant death of samples due to stress after sampling.34Some relevant studies have indicated that non-invasive or non-lethal sampling methods might affect the behavior or physiological state of animals in the short term.35,36 Through our long-term research, the results demonstrate that non-invasive and non-lethal sampling has no significant impact on fish. This is believed to be the first time that non-invasive or non-lethal sampling has been experimentally confirmed to have no long-term effects.
The application value of rare fish conservation and non-invasive sampling
The G. chilianensis as a representative species of plateau fish, holds significant importance for the conservation of plateau fish.14 Plateau fish have adapted to the unique plateau environment over a long period of evolution, developing distinct genetic traits. They predominantly inhabit the distinctive and fragile plateau aquatic ecosystem. Owing to the secluded and specific nature of the environment, many fish populations are small, rendering germplasm resources highly valuable.37 The loss of any individual can potentially have an irreversible impact on the entire population.38 Traditional invasive sampling methods, such as tissue dissection, directly result in the death of fish individuals, and the sampling process itself induces stress responses in the fish, potentially disrupting research on their normal physiological state. Noninvasive sampling avoids these additional stress factors, allowing for a more precise exploration of fish physiological characteristics. For certain rare plateau fishes like Schizothorax,39 each individual plays a crucial role in maintaining the population. Employing non-invasive sampling not only procures the necessary samples but also minimizes any impact on the samples, ensuring the fish can continue to survive and reproduce naturally, maintaining population stability. This, in turn, fosters sustainable fish research in the plateau region and aids in the protection of the local ecological environment.
Comparison of Non-invasive Sampling vs. Traditional Sampling Methods
Advances in sequencing technologies have enabled the development of efficient, low-cost genotyping methods, which are now critical tools in genetic studies of endangered fish species. Mucus, as a specialized physiological feature of fish, offers significant advantages for non-invasive DNA sampling. Surface mucus swabbing in G. chilianensis demonstrates simplicity, rapidity, and high efficiency, with minimal impact on sampled individuals. Immediate preservation of collected mucus in centrifuge tubes effectively prevents DNA contamination.
In contrast, traditional fin-clipping methods used by Zhang et al.14 yielded Cytb sequences (1100 bp) and D-loop sequences (750 bp), which align closely with the 1098bp Cytb and 696bp D-loop sequences obtained in this study via mucus swabbing. This confirms the feasibility of non-invasive mucus-based sampling for this species, achieving research objectives without compromising animal welfare.
Given the limited adoption of non-invasive methods in wild fish research, we advocate for their broader application in studies of germplasm resources and conservation. This approach maximizes the balance between animal protection and scientific requirements, offering an ethical framework for future biodiversity investigations.
Additionally, there are certain limitations in this study. The samples used are all from artificially bred populations, which differ from their wild counterparts. Meanwhile, the 70-day observation period was conducted entirely within the breeding environment, distinct from the wild environment where samples are returned after field collection. Hence, in subsequent research, we can utilize non-invasive sampling to conduct comparative genomics studies on the G. chilianensis populations distributed across various basins. This will help reveal the genetic relationships and evolutionary divergences among different populations, providing compelling evidence for the study of the biogeography and evolutionary history of the G. chilianensis. Furthermore, we can perform multiple non-invasive samplings of the G. chilianensis within the same basin to assess the impact of sampling methods on the fish through long-term dynamic monitoring.
Acknowledgments
This research is financially supported by the National Natural Science Foundation of China (32273140).
Authors’ Contribution
Data curation: Biyuan Liu (Equal), Qiqun Cheng (Equal). Formal Analysis: Biyuan Liu (Equal), Qiqun Cheng (Equal). Investigation: Biyuan Liu (Lead), Qiqun Cheng (Lead), Zhongyu Lou (Supporting), Di Peng (Supporting), Yong Qin (Supporting), Dan Song (Supporting). Methodology: Biyuan Liu (Lead), Qiqun Cheng (Lead), Zhongyu Lou (Supporting), Di Peng (Supporting), Yong Qin (Supporting), Dan Song (Supporting). Resources: Biyuan Liu (Lead), Qiqun Cheng (Lead), Zhongyu Lou (Supporting), Yong Qin (Supporting). Software: Biyuan Liu (Equal), Qiqun Cheng (Equal). Validation: Biyuan Liu (Equal), Qiqun Cheng (Equal), Zhongyu Lou (Supporting), Di Peng (Supporting), Yong Qin (Supporting), Dan Song (Supporting). Visualization: Biyuan Liu (Lead), Qiqun Cheng (Lead). Writing – original draft: Biyuan Liu (Lead). Writing – review & editing: Biyuan Liu (Lead), Qiqun Cheng (Lead), Zhongyu Lou (Supporting), Di Peng (Supporting). Conceptualization: Qiqun Cheng (Lead). Funding acquisition: Qiqun Cheng (Lead). Project administration: Qiqun Cheng (Lead). Supervision: Qiqun Cheng (Lead).
Competing Interest – COPE
The authors declare no conflicts of interest.
Ethical Conduct Approval – IACUC
All animal experiments in this study were conducted in accordance with the relevant national and international guidelines. Our project was approved by Institutional Animal Care & Use Committee (IACUC) of East China Sea Fisheries Research Institute.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All are available upon reasonable request.

_and_d-loop_region_(b)_in_cultured_.png)

_and_weight_(b)_in_experimental_group_and_control_group.png)

_and_mortality(b)_of_two_experimental_groups_and_control_group.png)
_and_d-loop_region_(b)_in_cultured_.png)

_and_weight_(b)_in_experimental_group_and_control_group.png)
_and_mortality(b)_of_two_experimental_groups_and_control_group.png)