INTRODUCTION
The Yuanshui River, a primary tributary of the Dongting Lake system in the middle reaches of the Yangtze River, is the largest river by water volume among the “Four Rivers” (Xiang, Zi, Yuan, Li) of Hunan Province. It originates from the Qingshui River in southeastern Guizhou Province. Flowing eastward, it acquires the name “Yuanshui River” after converging with the Wu Riverat Hongjiang City, Hunan Province. As the second-largest river in Hunan, the Yuanshui River’s main stem within the province stretches 568 km, draining a basin area of 51,066 km². Situated within a subtropical monsoon climate zone, the Yuanshui River basin experiences abundant annual rainfall and exhibits distinct geographical and hydrological characteristics. It encompasses diverse aquatic habitats, including rivers, wetlands, and reservoirs. The river course is conventionally divided into three sections: Upper Reaches, From its headwaters in the Qingshui River to Huaihua, Hunan, characterized by relatively swift flow; Middle Reaches, From Huaihua to Yuanling, featuring lower flow velocities and a landscape primarily interlaced with hills and valleys, with riverbeds predominantly composed of gravel; Lower Reaches, From Yuanling to Changde, where it converges into Dongting Lake, flowing mainly through hilly valley plains. The abundant water resources and diverse aquatic environments of the Yuanshui River provide varied habitats for aquatic organisms, supporting exceptionally rich fish resources.1 However, prior to the implementation of the decade-long fishing ban in the Yangtze River basin, frequent anthropogenic activities posed severe threats to fish survival. Overexploitation impeded long-lived fish species from reaching maturity, leading to pronounced miniaturization of fish populations and a significant decline in species diversity across river sections. The construction of dams and reservoirs obstructed natural river corridors, hindering fish migration. Consequently, the proportion of migratory fish species declined markedly, with diadromous species (migrating between rivers and the sea) becoming particularly scarce.2,3 To protect the aquatic ecosystem of the Yangtze River basin, the Ministry of Agriculture and Rural Affairs (MARA) of China instituted the Ten-year Fishing Ban in the Yangtze River. This comprehensive ban was formally implemented on January 1, 2021, covering key areas including the main stem of the Yangtze River, its major tributaries, large lakes connected to the river, and the Yangtze River estuary region. The ban aims to eliminate fishing pressure, improve the survival environment for fish within the Yangtze basin, and facilitate the recovery of its fish resources.4
Environmental DNA (eDNA) refers to the collective DNA fragments directly extracted from environmental samples (e.g., water, soil, air, ice cores), comprising a mixture of DNA from various organisms including microorganisms, animals, and plants. Environmental DNA metabarcoding analyzes fish DNA (e.g., from feces, skin, mucus) shed into the water column by collecting water samples, extracting DNA, and conducting sequencing.5 This non-invasive technique provides data on fish resources, including rare species, without capture, offering high sensitivity, environmental friendliness, and no harm to fish. It can serve as a monitoring technique for fish resources during the ten-year fishing ban period, complementing conventional fishing methods to obtain more comprehensive data on fish diversity. Within the context of this moratorium, this study investigates fish assemblages in the Yuanshui River using both eDNA metabarcoding and conventional fishing methods. It aims to assess fish diversity status during the ban, explore eDNA’s application for fish resource monitoring in the Yangtze River Basin, and provide insights for evaluating the moratorium’s effectiveness, conserving fish resources, and informing fisheries governance in the Yuanshui River and broader Yangtze Basin.
MATERIALS AND METHODS
FISH COMMUNITY SURVEY IN THE YUANSHUI RIVER USING eDNA TECHNOLOGY
WATER SAMPLE COLLECTION AND PROCESSING
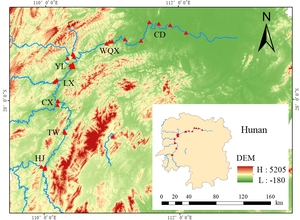
Environmental DNA (eDNA) water sampling was conducted in September 2023 across 38 sampling sites spanning from Potou Town, Changde City (lower reaches) to Hongjiang City (Figure 1). Sites were grouped into seven river sections: Changde, Wuqiangxi, Yuanling, Luxi, Chenxi, Tongwan, and Hongjiang. At each site, 2 liters of surface water were collected using a water sampler into sterilized polyethylene bottles. Gloves were replaced between each sample collection to prevent contamination. Within 5 hours of collection, water samples were vacuum-filtered through 0.45 μm pore-size hydrophilic mixed cellulose ester filters. All filtration equipment was disinfected between samples. Filters were placed into cryovials, flash-frozen in liquid nitrogen during transport, and subsequently stored at -80°C in the laboratory.
ENVIRONMENTAL DNA EXTRACTION, PCR AMPLIFICATION AND HIGH-THROUGHPUT SEQUENCING
Total DNA captured on filters was extracted using PowerWater DNA Isolation Kits. Extracted genomic DNA was verified via 1% agarose gel electrophoresis. PCR amplification targeted the freshwater fish 12S rRNA gene using universal primers Tele02_F (5’-AAACTCGTGCCAGCCACC-3’) and Tele02_R (5’-GGGTATCTAATCCCAGTTTG-3’). Reactions (20 μL) contained: 4 μL 5× FastPfu Buffer, 2 μL dNTPs (2.5 mmol/L), 0.8 μL each primer (5 μmol/L), 0.4 μL FastPfu Polymerase (2.5 U/μL), 0.2 μL BSA (0.8 μg/μL), 10 ng DNA template, ddH2O to 20 μL. Thermal cycling conditions: 95°C for 3 min (initial denaturation), 34 cycles of: 95°C for 30 s (denaturation), 58°C for 30 s (annealing), 72°C for 45 s (extension), 72°C for 10 min (final extension), Hold at 10°C. Amplified products were purified and recovered using 2% agarose gel electrophoresis. Environmental DNA Samples were sent to Shanghai Lingen Biotechnology Co., Ltd. for high-throughput sequencing on the Illumina NovaSeq 6000 platform.
FISH SPECIES IDENTIFICATION
Sequences obtained from high-throughput sequencing were processed using the DADA2 pipeline for primer removal, quality filtering, read merging, and chimera elimination to generate high-quality sequences. These processed sequences were then clustered into Operational Taxonomic Units (OTUs) at a 97% similarity threshold using QIIME2 software. For taxonomic assignment of each OTU, representative sequences were aligned and annotated against the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/) to achieve taxonomic classification at multiple levels (Kingdom, Phylum, Class, Order, Family, Genus, and Species). The annotation results were manually verified and cross-referenced with documented fish species data from the Yuanshui River, excluding non-fish sequences and marine fish species.1,6 The fish species composition at each sampling site was statistically analyzed. Taxonomic information was further refined by consulting authoritative references, including: Fauna Sinica, Osteichthyes, Cypriniformes (Volumes II & III); Fauna Sinica, Osteichthyes, Siluriformes; Hunan Fish Fauna; Key to the Freshwater Fishes of China.7–11
CLASSIFICATION OF FISH ECOLOGICAL GUILDS
Fish ecological guilds were classified according to three distinct schemes: (1) Based on ecological behavior,12 species were categorized into four groups: diadromous(between rivers and sea), river-lake migratory (between rivers and lakes), Lentic resident (preferring slow/still water), and Rheophilic (preferring flowing water); (2)Following trophic strategies,13 Yuanshui River fishes were grouped as omnivorous, carnivorous, and herbivorous; (3) According to vertical stratification,14 species were classified as upper water column, lower water column, and demersal. This tripartite classification system provides a comprehensive functional characterization of the fish assemblage, integrating migratory patterns, feeding ecology, and habitat partitioning within the aquatic ecosystem.
FISH COMMUNITY SURVEY IN THE YUANSHUI RIVER USING CONVENTIONAL FISHING GEAR
Concurrently with eDNA sampling, fish resources were sampled in four river sections (Changde, Yuanling, Chenxi, Hongjiang) of the Yuanshui River basin using conventional fishing gear combined with market surveys. Primary gear types included gillnets (mesh sizes: 100 mm, 70 mm, 50 mm, 30 mm, 10 mm) and fyke nets. Two gillnets of each mesh size (each 100 m long) and fyke nets (total length 200 m) were deployed daily in the afternoon and retrieved the following morning, resulting in an approximate 10-hour soak time per set. Captured fish were processed immediately: identified to species, photographed, measured for body length and weight. Specimens requiring further identification were frozen for transport to the laboratory. This survey was approved by the Hunan Provincial Department of Agriculture and Rural Affairs and coordinated with local fisheries authorities and fishers.
DATA STATISTICS AND PROCESSING
Alpha diversity indices (e.g., Shannon, Simpson, Chao1, and Pielou indices) for fish communities in each river section, derived from environmental DNA (eDNA) data, were calculated using R Studio. Additionally, diversity indices obtained via conventional methods were computed using the following formulas, enabling a comprehensive assessment of fish diversity across sampling sites.
Shannon-Wiener Diversity Index (H’): H’ =
Simpson Diversity Index: GS =
Pielou Evenness Index (E): E =
RESULTS
FISH SPECIES COMPOSITION BASED ON ENVIRONMENTAL DNA
Clustering analysis and taxonomic annotation identified a total of 94 fish species, belonging to 10 orders, 17 families, and 57 genera. Cypriniformes was the dominant order (65 species, 69.15%), followed by Siluriformes (11 species, 11.70%), Perciformes and Gobiiformes (5 species each, collectively 10.64%), and Anabantiformes (3 species, 3.19%). The remaining orders each contained only one species (collectively 5.32%). At the family level, Cyprinidae (Cypriniformes) was the most diverse group (54 species, 57.45%), followed by Bagridae (Siluriformes; 8 species, 8.51%) and Botiidae (Cypriniformes; 6 species, 6.38%). Serranidae (Perciformes) comprised 5 species (5.32%), and Gobiidae (Gobiiformes) contained 4 species (4.26%). Cobitidae accounted for 3 species (3.19%), while Nemacheilidae, Siluridae, and Osphronemidae each had 2 species (collectively 6.38%). Eight families (Channidae, Engraulidae, Salangidae, Hemiramphidae, Mastacembelidae, Poeciliidae, Eleotridae, and Ictaluridae) were represented by a single species each (Table 1).
Among the 94 detected fish species in the Yuanshui River: Endemic to the Yangtze Basin (12 species): Culter oxycephaloides, Hemiculterella sauvagei, Megalobrama amblycephala, Megalobrama pellegrini, Squalidus wolterstorffi, Saurogobio gymnocheilus, Acheilognathus hypselonotus, Acrossocheilus jishouensis, Decorus tungting, Cobitis macrostigma, Leptobotia taeniops. Non-native Species (3 species): Luciobarbus capito, Ictalurus punctatus, Gambusia affinis. Hunan Province Key Protected Wildlife (11 species): Coilia nasus, Ochetobius elongatus, Coreius heterodon, Saurogobio dumerili, Acheilognathus hypselonotus, decorus tungting, Parabotia kiangsiensis, Leiocassis longirostris, Macropodus chinensis, Macropodus opercularis, Siniperca roulei.
FISH ECOLOGICAL GUILDS
Based on ecological habits,12 Yuanshui River fishes identified via eDNA were categorized into four types: (1) Diadromous : Coilia nasus(1 species); (2) River-lake migratory: 14 species (14.89%), including Ochetobius elongatus, Elopichthys bambusa, and the Chinese carps (black, grass, silver, bighead), etc.; (3) Lentic resident: 53 species (56.38%), including three Culter spp., five Acheilognathinae species, four Pelteobagrus spp., Cyprinus carpio, and Carassius auratus, etc; (4) Rheophilic: 26 species (27.66%), including Squalidus wolterstorffi, Leptobotia taeniops, and Leptobotia guilinensis, etc.
Based on feeding habits,13 Yuanshui River fishes were categorized into three trophic guilds: (1) Omnivorous (60 species, 63.83%): Includes most Cobitidae and Cyprininae species, Pelteobagrus spp., Pseudobagrus spp., etc.; (2) Carnivorous (28 species, 29.79%): Includes five Siniperca spp., three Culter spp., two Macropodus spp., Odontobutis obscura, Rhinogobius giurinus, etc.; (3) Herbivorous (6 species, 6.38%): Includes Ctenopharyngodon idellus (grass carp), Xenocypris davidi, Megalobrama spp, etc.
Based on habitat preference in the water column,14 Yuanshui River fishes were categorized into three groups: (1) Upper water column (34 species, 36.17%): Includes Hemiculter leucisculus, Culter alburnus, Siniperca kneri, etc; (2) Lower water column (17 species, 18.09%): Includes Xenocypris davidi, Silurus meridionalis, Macropodus chinensis, etc.; (3) Demersal (43 species, 45.74%): Includes Cobitidae, Botiidae, Bagridae, and other bottom-dwelling species.
SPATIAL DISTRIBUTION OF FISH DIVERSITY
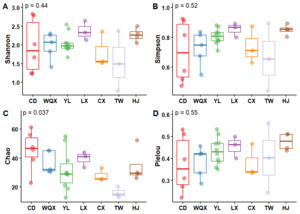
Analysis of fish species distribution and diversity across 38 sampling sites in 7 sections of the Yuanshui River revealed: Species richness of Changde (70 species), Wuliangxi Reservoir (56), Yuanling (71), Luxi (56), Chenxi (44), Tongwan Town (25), Hongjiang (49); Average diversity indices (range) of Shannon-Wiener: 1.55 - 2.37, Simpson: 0.66 - 0.85, Chao1 richness: 16.17 - 31.43, Pielou evenness: 0.37 - 0.48 (Figure 2). The Shannon-Wiener and Simpson indices exhibited similar spatial patterns, peaking in Luxi and lowest in Tongwan Town. The highest Chao1 richness occurred in Changde (45.35), while the lowest was in Tongwan Town (16.17). Pielou evenness was highest in Hongjiang (0.48) and lowest in Changde (0.37). Although Yuanling had the highest species richness (71), its average Shannon index (2.04) was not the highest, likely due to the relatively low per-site species richness and uneven distribution across its 10 sampling sites.
COMPARISON OF FISH SPECIES DIVERSITY USING eDNA TECHNOLOGY VERSUS CONVENTIONAL FISHING GEAR
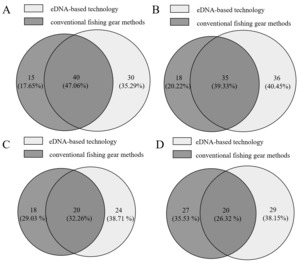
To facilitate the comparison of environmental DNA (eDNA) metabarcoding and conventional fishing gear-based surveys in assessing fish biodiversity in the Yuanshui River, we analyzed datasets from identical river sections in Changde, Yuanling, Chenxi, and Hongjiang. The results revealed significant differences in species detection rates between methodologies. eDNA metabarcoding identified the following fish species richness: 70 species in Changde, 71 species in Yuanling, 44 species in Chenxi, and 49 species in Hongjiang. In contrast, conventional surveys detected substantially fewer species: 55 species in Changde, 53 species in Yuanling, 38 species in Chenxi, and 47 species in Hongjiang. As shown in Figure 3, in the Changde section of the Yuan River, 85 total species were detected, 40 shared species, and eDNA detected 15 additional species; in the Yuanling section, 89 total species were detected, 35 shared species, and eDNA detected 18 additional species; in the Chenxi section: 62 total species were detected, 20 shared species, eDNA detected 6 additional species; Hongjiang section: 76 total species detected, 20 shared species, eDNA detected 2 additional species.
A comparison of Shannon diversity (H’) and Pielou evenness (E) indices between eDNA metabarcoding and conventional capture-based methods across the four river sections (Changde, Yuanling, Chenxi, Hongjiang) revealed significantly higher values for both indices obtained via conventional methods (Table 2).
DISCUSSION
ANALYSIS OF FISH SPECIES DIVERSITY IN THE YUANSHUI RIVER USING eDNA TECHNOLOGY AND CONVENTIONAL FISHING GEAR
This study employed environmental DNA (eDNA) technology to detect fish species at 38 sampling sites across 7 sections of the Yuanshui River. A total of 94 fish species were identified, belonging to 10 orders, 17 families, and 57 genera. Cypriniformes dominated (65 species, 69.15%), followed by Siluriformes (11 species, 11.70%), Perciformes and Gobiiformes (5 species each, 5.32%), Anabantiformes (3 species, 3.19%), with other orders represented by single species. During 2022–2023, conventional fishing gear surveys detected 120 fish species. Cypriniformes remained predominant (85 species, 70.83%), followed by Siluriformes (17 species, 14.17%), Perciformes (9 species, 7.50%), Gobiiformes (4 species, 3.33%), Anabantiformes (2 species, 1.67%), and singleton species in other orders.15
Comparison of fish species richness revealed that environmental DNA (eDNA) technology detected significantly fewer species than conventional fishing gear methods. This discrepancy primarily stems from differences in sampling duration and frequency: conventional methods aggregated data from biannual surveys (spring-summer and autumn-winter) conducted over 2022–2023, with each survey spanning 10 days; eDNA sampling was completed once in September 2023. Given the spatiotemporal dynamics of fish distribution, a single eDNA sampling event inherently limits comprehensive species detection in the Yuanshui River. Fish species composition detected by environmental DNA (eDNA) technology and conventional fishing gear exhibited high congruence, with Cypriniformes dominant (69.15% vs. 70.83%), followed by Siluriformes (11.70% vs. 14.17%), Perciformes (5.32% vs. 7.50%), Gobiiformes (5.32% vs. 3.33%), and Anabantiformes (3.19% vs. 1.67%). Remaining orders contained singleton species. This taxonomic structure aligns with documented ichthyofauna characteristics in the middle Yangtze River.1,16 Additionally, eDNA detected 12 endemic Yangtze species, 10 of which (Culter oxycephaloides, Megalobrama amblycephala, M. pellegrini, Squalidus wolterstorffi, Saurogobio gymnocheilus, Acheilognathus hypselonatus, Acrossocheilus jishouensis, Sinilabeo decorus, Cobitis macrostigma, Leptobotia taeniops) were confirmed by conventional methods.15 In conclusion, eDNA achieves comparable detection efficacy to conventional sampling for fish community assessment.
Comparative analysis of fish communities across four Yuanshui River sections (Changde, Yuanling, Chenxi, Hongjiang) revealed distinct spatial patterns in detection efficacy between environmental DNA (eDNA) technology and conventional fishing gear. Species richness values were: Changde (eDNA:70, conventional :55), Yuanling (71:53), Chenxi (44:38), and Hongjiang (49:47). Shared species counts were Changde:40, Yuanling:35, Chenxi:20 and Hongjiang:20 (Figure 3). Both methodologies demonstrated significantly higher species richness and shared species detection in the lentic-influenced Changde and Yuanling sections compared to the lotic Chenxi and Hongjiang reaches. This spatial heterogeneity is attributed to the lentic characteristics of Changde/Yuanling-featuring low-flow hydrology and broad channel morphology that enhance eDNA source accumulation (mucus, excreta) and conventional capture efficiency. Conversely, the mid-upper reaches (Chenxi/ Hongjiang) exhibit faster-flowing lotic conditions that reduce detection of lacustrine-adapted species and accelerate eDNA dispersal, resulting in diminished overall detection efficacy for both methodologies.
Table 2 indicates that Shannon diversity indices and Pielou evenness indices obtained via conventional methods consistently exceeded those derived from environmental DNA (eDNA) technology across all four river sections. This discrepancy may be attributed to:(1) Spatiotemporal sampling limitations: eDNA sampling points failed to capture benthic sediments containing fish scales, mucus, and excreta17; Incomplete coverage of transient fish activity hotspots. (2) eDNA degradation drivers include Microbial processing, Temperature-dependent decay kinetics, Shear stress from hydrological forcing. These factors collectively reduced detection sensitivity at individual sampling points, ultimately depressing section-wide diversity metrics below conventional survey values.
APPLICATION OF eDNA TECHNOLOGY IN FISH DETECTION IN THE YANGTZE RIVER BASIN
Environmental DNA (eDNA) technology serves as an emerging biomonitoring tool for aquatic ecosystems. Current research demonstrates its distinct advantages over conventional fishery surveys for fish species detection: (1) Enhanced cost-effectiveness through reduced temporal and labor expenditures18; (2)Non-invasiveness that preserves habitat integrity and prevents bycatch mortality; (3)Reduced taxonomic subjectivity in species identification19; and (4)Superior detection sensitivity enabling effective monitoring of elusive taxa including rare endemics, invasive species, and small-bodied fishes with low population densities or challenging sampling requirements.20
Current applications of environmental DNA (eDNA) technology in the Yangtze River Basin primarily focus on assessing fish biodiversity. Li et al.21integrated eDNA metabarcoding with conventional early-life-stage surveys in the Nanjing section, detecting 39 fish species. This approach addressed the limitation of conventional methods in capturing mesopelagic early-life stages, significantly enhancing sampling efficiency and species detection rates. Similarly, Zhang et al.22identified 37 species in the Jiangjin section through eDNA analysis, including rare endemic species such as Procypris rabaudi and Acipenser dabryanus that are typically undetectable by conventional methods. Wu et al.23 employed high-quality eDNA extraction to study Neophocaena asiaeorientalis distribution, detecting eDNA signals at all 8 sites with visual observations and at 3 of 10 sites without sightings. This confirms the feasibility of eDNA metabarcoding for aquatic ecological monitoring, demonstrating both detection reliability and enhanced sensitivity compared to conventional survey methods.
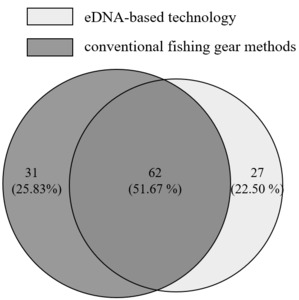
Comprehensive analysis of fish species detection across four Yuanshui River sections revealed a total of 89 species identified via environmental DNA (eDNA) technology versus 93 species through conventional methods. Concurrent detection by both methodologies recorded 62 shared species (Figure 4), representing 69.66% of eDNA-detected taxa and 66.67% of conventionally surveyed species. This high methodological congruence demonstrates the reliability of eDNA approaches for fish biodiversity assessments, validating their application in riverine ecological studies.
Analysis of methodological detection discrepancies revealed that among the 89 species detected by environmental DNA (eDNA) across four river sections, 27 were undetected by conventional fishing gear. These included demersal species (Neosalanx taihuensis, Spinibarbus hollandi, Cobitis macrostigma, Parabotia lijiangensis, Leptobotia guilinensis) and small-bodied fish (Hemiculterella sauvagi, Toxabramis swinhonis, Microphysogobio fukiensis, Saurogobio punctatus, Hyporhamphus intermedius), demonstrating eDNA’s superior detection efficacy for these ecological guilds. Regarding threatened species, both methods detected IUCN Critically Endangered (CR) species Ochetobius elongatus and Pseudobagrus eupogon, along with Hunan Province-protected species (O. elongatus, Coreius heterodon, Acheilognathus hypselonatus, Leiocassis longirostris, Siniperca roulei, Macropodus ocellatus, M.erythropterus, decorus tungting). However, eDNA exclusively detected three provincially protected taxa: Coilia nasus, Saurogobio dumerili, and Parabotia kiangsiensis. Figure 4 indicates 31 species captured conventionally were undetected by eDNA. Contributing factors beyond the single September 2023 sampling include: (1)"False negatives"from: a) Primer limitations, despite using optimal freshwater fish primers (12S rRNA), reduced taxonomic resolution occurs in closely related taxa due to minimal genetic divergence in standard barcodes. b) Database constraints, incomplete reference sequences limit species annotation accuracy, particularly for regional endemics.24,25 (2) Temporal sampling bias failing to capture seasonal occurrence patterns. These operational constraints highlight persistent challenges in eDNA-based biodiversity monitoring despite its demonstrated advantages.
In conclusion, this study demonstrates that eDNA-based assessments of fish diversity in the Yuanshui River yield scientifically robust results. Nevertheless, the application of eDNA technology for freshwater fish surveys in China’s inland waters faces persistent methodological constraints. Until regional ichthyofauna databases are comprehensively curated through conventional fisheries surveys, eDNA methodology should be implemented as a complementary approach rather than a complete replacement for conventional gear-based sampling. Integrating multi-method data within a unified analytical framework enhances the accuracy and comprehensiveness of fish biodiversity assessments in target aquatic ecosystems.
ACKNOWLEDGMENTS
Foundation Project supported this work: National Key R&D Program of China (2022YFD2400102); National Natural Science Foundation of China (Grant No.31572619)
AUTHORS’ CONTRIBUTION
Conceptualization: Liangguo Liu (Lead), Tangsi Zhang (Equal); Methodology: Liangguo Liu (Equal), Tangsi Zhang (Equal), Congqiang Luo (Equal), Xiao Xie (Equal), Yushuang Luo(Equal), Bolan Song(Equal), Jinlong Wang(Equal); Formal Analysis: Liangguo Liu (Equal), Tangsi Zhang (Equal); Investigation: Liangguo Liu (Equal), Tangsi Zhang(Equal); Writing–original draft: Tangsi Zhang (Equal), Liangguo Liu (Equal); Funding acquisition: Liangguo Liu (Equal), Congqiang Luo (Equal); Resources: Liangguo Liu (Equal), Congqiang Luo (Equal); Supervision: Liangguo Liu (Equal), Tangsi Zhang (Equal); Writing – review & editing: Liangguo Liu(Lead), Tangsi Zhang (Equal).
COMPETING INTEREST–COPE
No competing interests were disclosed.
ETHICAL CONDUCT APPROVAL–IACUC
The experimental protocols adhered to the ethical guidelines set forth by the Committee on the Ethics of Animal Experiments of Hunan University of Arts and Science, China.
INFORMED CONSENT STATEMENT
All authors and institutions have confirmed this manuscript for publication.
DATA AVAILABILITY STATEMENT
The data that has been used is confidential.