Introduction
The aquaculture industry plays a vital role in global food security and protein supply. However, the sustainable development of this industry is constantly challenged by issues such as infectious diseases and suboptimal growth performance, resulting in significant economic losses.1,2 The Chinese soft-shelled turtle, Pelodiscus sinensis, represents an economically important cultured species in China, highly valued for its nutritional and medicinal properties.3 According to the China Fishery Statistical Yearbook 2025, China’s annual turtle production reached a level of over 540,000 metric tons in 2024. However, the intensive cultivation of P. sinensis is frequently hindered by a low growth rate and high disease incidence, underscoring the urgent need for genetic improvement and the development of effective molecular breeding strategies.4 Addressing these complex phenotypic problems requires a fundamental understanding of the underlying molecular mechanisms that concurrently govern immune competence and somatic growth in this species.
The growth and immune responses in vertebrates are intricately regulated by complex signaling networks. Among these, the JAK/STAT signaling pathway is a central mediator of cytokine signaling, critically involved in immunity, development, and cellular homeostasis.5 The suppressor of cytokine signaling (SOCS) family proteins, particularly SOCS1, act as a negative regulator of JAK/STAT signaling pathway, involve in the regulation of immune, developmental, and metabolic homeostasis processes in vertebrates.6–8 In recent years, the functional analysis of SOCS1 gene in mammals and fish revealed that, in addition to modulating inflammatory response and antiviral immunity, SOCS1 also plays a role in the development of individuals through regulating the growth hormone (GH)-insulin-like growth factor (IGF) axis.9,10 Additionally, SOCS1 gene knockout in zebrafish (Danio rerio) displayed decreased insulin signaling sensitivity, hepatic steatosis, and sustained hyperactivation of the growth hormone signaling pathway.11 The SOCS family genes are ancient and conserved across vertebrates, yet their functional diversification and regulatory mechanisms in different vertebrate lineages remain incompletely understood. Specifically, the role of SOCS1 in non-avian reptiles is virtually unexplored, creating a significant gap in our comprehension of the evolutionary history of the JAK/STAT pathway and its regulation.
Therefore, this study aims to bridge this gap by conducting the first comprehensive investigation of the SOCS1 gene in P. sinensis. We reported the first cloning and bioinformatics analysis of SOCS1 gene in P. sinensis. The coding sequence and protein characteristics of SOCS1 were identified. The tissue-specific expression profile and response to sex steroid hormone were investigated. Collectively, these findings provide a fundamental molecular resource for understanding the biological functions of SOCS1 and advancing the genetic breeding of P. sinensis.
Materials and Methods
Experimental Materials
All turtles used in the experiment were purchased from the breeding base of Hezhou Soft-shelled Turtle Industrial Park in Changde City, Hunan Province. These turtles were in normal development and good health. Tissue samples, including the heart, liver, spleen, kidney, testis, ovary, intestine, muscle, and brain of adult turtles, were collected. After being rapidly frozen in liquid nitrogen, the samples were stored in a - 80°C refrigerator for future use.
A total of 150 juvenile turtles (average body mass: 35.42 ± 14.25 g) were randomly assigned to three groups: a methyltestosterone (MT) group, a 17β-estradiol (E2) group, and a control group, with 50 individuals per group. The hormone dosage (10 mg/kg for both E2 and MT) was selected based on previous studies on sex steroid hormone responses in P. sinensis, which demonstrated effective gene induction within this range while minimizing adverse effects on survival.12–14 For administration, 15 mg of E2 or MT was first dissolved in 0.5 mL of absolute ethanol and then diluted with 4.5 mL of olive oil. The control group received the vehicle solution (0.5 mL absolute ethanol in 4.5 mL olive oil). After one week of fasting with free access to water, each turtle was intramuscularly injected in the hind limb thigh with approximately 0.1 mL of the corresponding solution. Gonad samples were collected at 0-, 6-, 12-, 24-, and 48-hours post-injection, with 10 individuals (5 males and 5 females) sampled at each time point. All samples were immediately frozen in liquid nitrogen and stored at –80°C for subsequent analysis.
Cloning of the P. sinensis SOCS1 Gene
Based on the predicted sequence of the P. sinensis SOCS1 gene, specific amplification primers were designed using Primer Premier 6.0 software (Table 1). A core fragment was first amplified using a mixed-tissue cDNA pool as the template. Gene-specific primers for 5’- and 3’-RACE were then designed based on this fragment. The SMARTer® RACE 5’/3’ Kit (TaKaRa) was used, and the rapid amplification of cDNA ends was carried out strictly following the operation procedures in the instruction manual. All PCR products were detected by 1.5% agarose gel electrophoresis. After gel-cutting and recovery, they were ligated with the Peasy-T5 Zero vector and transformed. Positive clones were identified through blue-white screening and colony PCR. Finally, the positive bacterial solutions were selected and sent to a sequencing company for sequence determination.
Bioinformatics Analysis of the P. sinensis SOCS1 Gene
The DNAMAN software was used to splice the sequences to obtain the full-length cDNA of the target gene. The ORF Finder on NCBI was utilized to search for the open reading frame (ORF) of the target gene and predict its deduced amino acid sequence. The amino acid sequence of P. sinensis SOCS1 was subjected to a homology comparison with the amino acid sequences of other species in NCBI through local Blast. The STRING database was used to construct the protein-protein interaction network of SOCS1. A phylogenetic tree was constructed using the Neighbor-joining (NJ) method in MEGA 11.0 software. Through the NCBI database, the upstream and downstream gene sequences of the SOCS1 gene were obtained from the genomic data of P. sinensis and other species respectively, for the collinearity analysis of SOCS1 and its surrounding genes.
Tissue Expression Analysis of the P. sinensis SOCS1 Gene
In this study, real-time quantitative PCR (qPCR) was employed to investigate the expression profile of the SOCS1 gene in various tissues of P. sinensis. RNA samples from nine tissues, including the liver, kidney, spleen, intestine, pituitary, heart, muscle, lung, and gonads, were selected. After quality inspection (A260/A280 ratio of 1.8-2.0) and concentration determination, reverse transcription was carried out using the PrimeScript™ RT reagent Kit (TaKaRa) to synthesize cDNA.
qPCR amplification was performed on a LightCycler® 480 II system (Roche). The quantitative PCR reaction system (10 µL) consisted of: 1 µL of cDNA template (diluted to 50 ng/µL), 0.25 µL each of forward and reverse primers (10 µM), 5 µL of 2×Rapid Taq Master Mix (Vazyme), and 3.5 µL of nuclease-free water. The reaction procedure was as follows: pre-denaturation at 95℃ for 5 min; then 40 cycles of amplification (95℃ for 15 s, 58℃ for 15 s, 72℃ for 15 s, and the fluorescence signal was collected at the extension phase of 72℃). To verify amplification specificity, a melting curve analysis was performed by heating from 65°C to 97°C at a rate of 0.1°C/s with continuous fluorescence monitoring
Gene expression analysis was conducted using the 2^(–ΔΔCt) method, with GAPDH serving as the internal reference gene for standardization. Statistical analysis was performed using SPSS 22.0 with one-way ANOVA, where a P < 0.05 was considered statistically significant. Data are presented as mean ± standard deviation (Mean ± SD), and graphs were generated using GraphPad Prism 8.0.2.
Results
Molecular Characterization of P. sinensis SOCS1
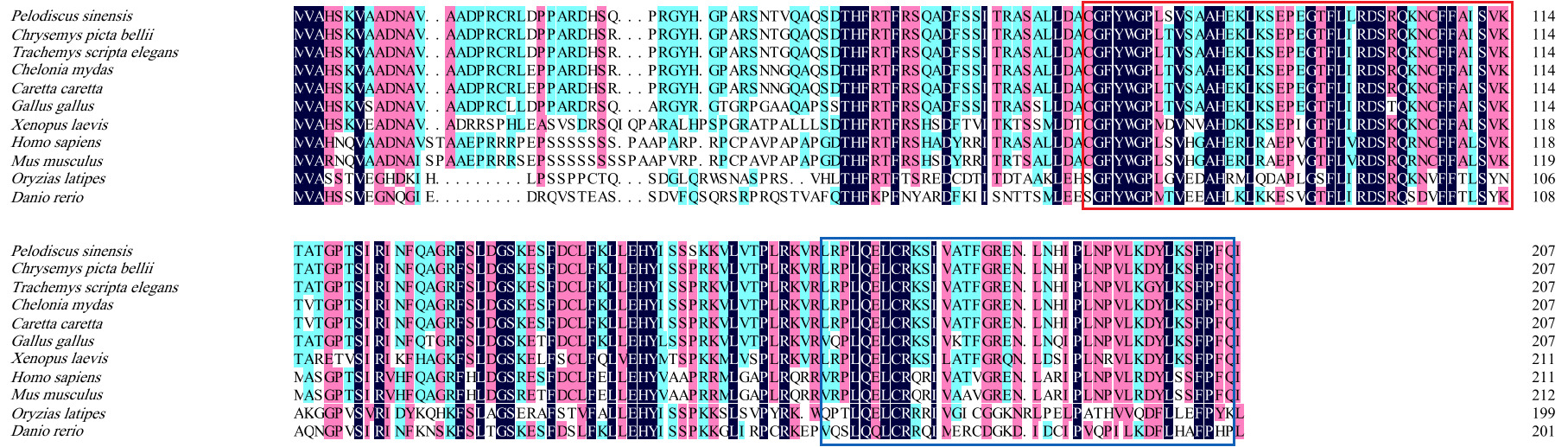
The full-length sequence of the P. sinensis SOCS1 gene is 2782 bp, containing a complete coding sequence. Bioinformatics analysis revealed that the ORF of this gene is 621 bp in length, encoding a polypeptide of 207 amino acid residues. The predicted protein has a molecular weight of 23.5 kDa and a theoretical isoelectric point of 8.72. Conserved domain analysis identified characteristic SOCS family motifs, including an SH2 domain (amino acids 73–156) and a C-terminal SOCS box (amino acids 167–203) (Figure 1).
Evolutionary Conservation and Homology Analysis
Sequence homology analysis via Blastp revealed a clear phylogenetic pattern in SOCS1 conservation across vertebrates (Table 2). The highest homology with P. sinensis was observed in reptiles, ranging from 95.65% (Chelonia mydas and Caretta caretta) to 97.10% (Chrysemys picta bellii). This was followed by 88.89% in the bird Gallus gallus. As evolutionary distance increased, homology declined to 68.25% in the amphibian Xenopus laevis; 63.98% and 61.79% in the mammals Homo sapiens and Mus musculus; and to the lowest values of 50.00% and 46.86% in the fish Oryzias latipes and Danio rerio, respectively. This correlation between sequence divergence and taxonomy suggests high functional conservation of SOCS1 during vertebrate evolution. Notably, the SH2 and SOCS box domains exhibited highly similarity across all species, indicating they have undergone strong purifying selection (Figure 2).
Phylogenetic Relationships of SOCS1
To elucidate the evolutionary relationships of SOCS1, a phylogenetic tree was constructed using the Neighbor-Joining method in MEGA 11.0. The resulting tree showed that vertebrates clustered monophyletically into distinct groups corresponding to reptiles, birds, mammals, amphibians, and fish (Figure 3). P. sinensis exhibited the closest relationship with C. picta bellii, and collectively, reptiles formed a sister clade to birds, reflecting their shared evolutionary ancestry. The phylogenetic distance to fish was the largest, which aligns with the amino acid sequence homology analysis and underscores the molecular divergence between these lineages.
Protein-Protein Interaction Network Prediction
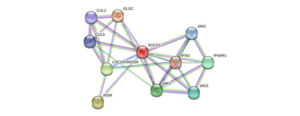
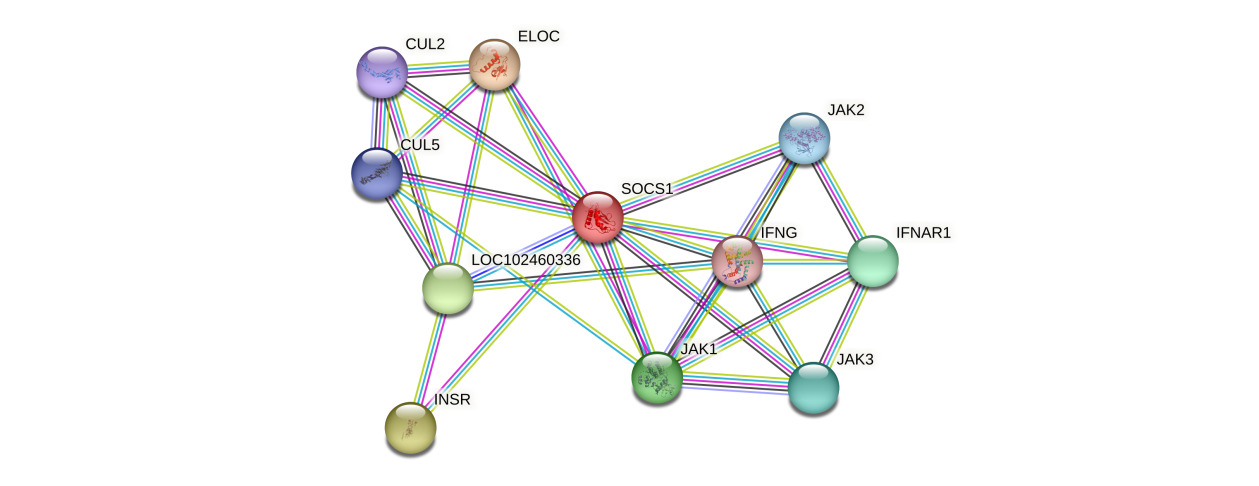
Protein-protein interaction network analysis based on the STRING database (version 11.5) indicated that the P. sinensis SOCS1 protein forms a complex regulatory network with 10 core interacting proteins. These include cytokine signaling molecules (IFNG, IFNAR1), members of the Janus kinase family (JAK1/2/3), components of the ubiquitin-ligase (ELOC, CUL2/5), the insulin receptor (INSR), and the uncharacterized protein LOC102460336 (Figure 4). The topological features of the network showed that SOCS1 had the highest node degree, occupying a pivotal position in the entire interaction network. Members of the JAK family not only maintained strong interactions with SOCS1 but also formed a tight functional module among themselves, jointly constituting the regulatory core of the JAK-STAT signaling pathway. This network exhibited distinct functional modularity features: the signal transduction module (JAKs, IFNG, IFNAR1) was responsible for cytokine signal transmission; the protein degradation module (CULs, ELOC) mediated the ubiquitination of target proteins; and the metabolic regulation module (INSR) was involved in energy metabolism balance. Notably, the high-score interaction between the uncharacterized protein LOC102460336 and SOCS1 implied the possible existence of new regulatory mechanisms. These findings provided important clues for in-depth analysis of the functional mode of P. sinensis SOCS1, which integrates SH2 domain-mediated signal suppression-SOCS box-dependent protein degradation-metabolic receptor regulation. Meanwhile, it also highlighted the central position of SOCS1 in the immunometabolism regulation network.
Conserved Synteny and Lineage-Specific Insertions of the SOCS1 Gene in P. sinensis
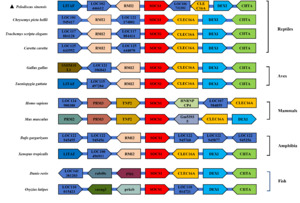
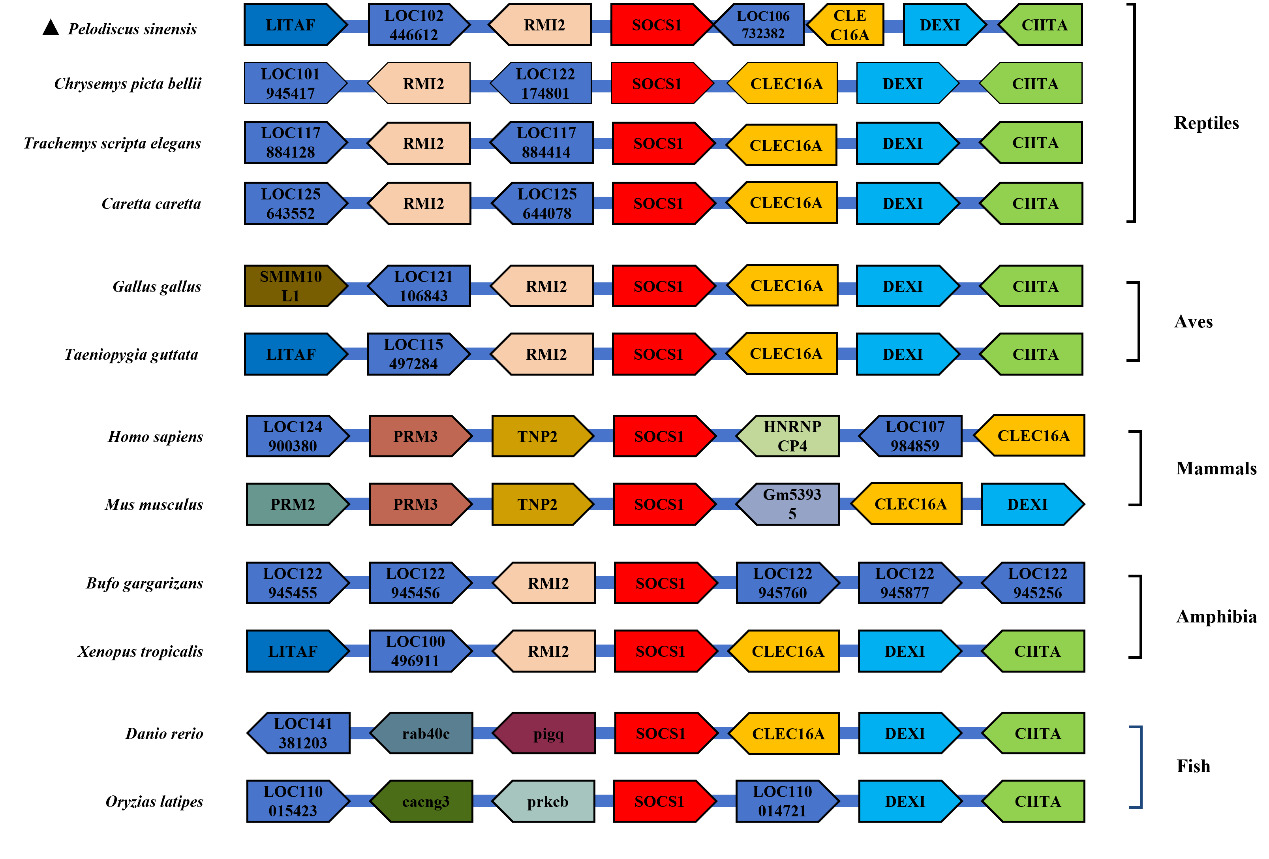
To conduct an in-depth study of the collinearity characteristics of the SOCS1 gene, based on the SOCS1 genomic sequence obtained from the NCBI database, a collinearity diagram of the SOCS1 gene in P. sinensis and that in other species, along with its seven adjacent genes upstream and downstream, was drawn. As shown in Figure 5, other adjacent genes of the SOCS1 gene, such as CLEC16A, DEXI, and CIITA, exhibited collinearity in multiple species. In closely related species of P. sinensis, including C. picta bellii, T. scripta elegans, and C. caretta, the order and relative positions of these genes were relatively similar. However, in P. sinensis, other genes were inserted, which might be attributed to gene expansion or loss on the chromosomes of different species during biological evolution.
Tissue-Specific and Sexually Dimorphic Expression of SOCS1
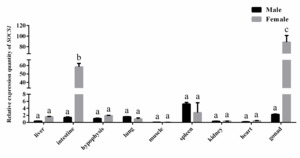
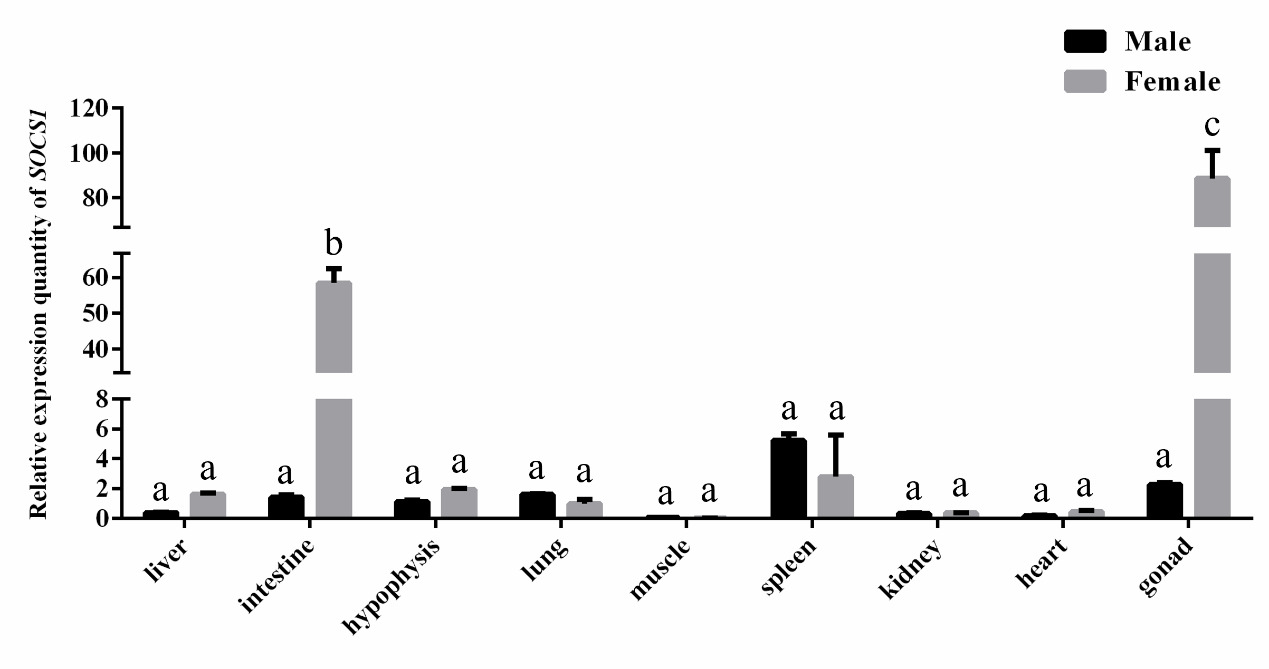
Tissue-specific and sexually dimorphic expression of the SOCS1 gene was identified in P. sinensis via qPCR analysis (Figure 6). The expression level of SOCS1 was relatively higher in gonads, intestine, and spleen than that in other tested tissues, indicating that SOCS1 might be involved in physiological regulation of these tissues. Furthermore, comparative analysis between sexes revealed that SOCS1 expression was significantly upregulated in the intestine and gonads of females compared to males (P < 0.05). This sexually dimorphic expression pattern suggests that SOCS1 may be involved in sex-specific physiological regulation in P. sinensis.
Hormonal Regulation of SOCS1 in Gonads
qPCR analysis revealed a sexually dimorphic pattern in the hormonal regulation of gonadal SOCS1. In males, SOCS1 expression remained unaltered by either E2 or MT exposure, exhibiting only non-significant fluctuations (P > 0.05). In females, however, the two hormones elicited strikingly opposite effects: E2 induced a significant transient upregulation, characterized by a “bell-shaped” expression curve that peaked at 12 h (P < 0.05). In contrast, MT significantly downregulated SOCS1 expression at the same time point (12 h, P < 0.05) (Figure 7).
Discussion
This study presents the first systematic analysis of the SOCS1 gene in P. sinensis, elucidating its molecular characterization, evolutionary trajectory, and putative physiological functions in reptiles. We successfully cloned the SOCS1 cDNA, which is 2,782 bp in full length and contains a 621 bp open reading frame encoding a 207-amino acid protein. The predicted protein possesses hallmark domains of the vertebrate SOCS family, including a highly conserved SH2 domain (aa 73-156) and a SOCS box (aa 167-203), suggesting its potential role in the JAK-STAT signaling pathway.15 Moreover, the molecular weight (23.5 kDa) and isoelectric point (8.72) of the SOCS1 protein in the P. sinensis were almost identical to those of other turtle species. This high similarity in basic physicochemical properties, coupled with the conserved domain architecture described above, suggests a potential conservation of protein structure and, by extension, biological function among turtles.
Phylogenetic analysis revealed that the sequence similarity of P. sinensis SOCS1 closely reflects established taxonomic relationships.16 It shared the highest identity (>95%) with C. picta bellii, followed by ~88% in birds and ~62% in mammals. Notably, the SH2 domain and SOCS box were highly conserved across all compared species, likely due to strong purifying selection during vertebrate evolution. This finding provides new evidence supporting the functional conservation of SOCS family proteins in vertebrates.
Protein-protein interaction network analysis identified P. sinensis SOCS1 as a central hub with strong predicted interactions, including members of the JAK kinase family, components of the ubiquitin-ligase complex, and the insulin receptor.17 This supports a model wherein SOCS1 may integrate SH2 domain-mediated signal inhibition with SOCS box-dependent protein degradation and metabolic receptor regulation. Furthermore, a high-confidence interaction with the uncharacterized protein LOC102460336 was detected, suggesting a potential novel regulatory mechanism specific to turtles, which offers a valuable direction for future research.
The study systematically elucidated the expression characteristics of the P. sinensis SOCS1 gene and its sexually dimorphic expression pattern for the first time. Tissue-specific expression analysis showed that SOCS1 was significantly highly expressed in the gonads and intestine of female individuals (P < 0.05). Although previous studies have suggested that SOCS1 is involved in physiological processes such as immune regulation, growth and development, and metabolic balance, its role in gonadal development has not been previously reported.18 Based on the regulatory mechanism of the JAK-STAT signaling pathway, we speculate that the high expression of SOCS1 in the female gonads of P. sinensis may participate in the dynamic balance of follicular development and vitellogenesis by finely regulating the activity of the JAK-STAT signaling pathway. Crucially, a key aspect of maintaining a healthy ovarian microenvironment for successful folliculogenesis is the establishment of immune privilege, which protects developing oocytes from immune-mediated damage. Meanwhile, it may mediate the signal transduction of reproductive cycle-related hormones (such as estrogen) and provide a unique immune-privileged microenvironment for oocytes to ensure the normal development.19–21 Notably, the sexually dimorphic expression pattern observed in the intestine may reflect the special nutritional and metabolic demands of female individuals during the reproductive period.22 This tissue-specific expression regulation may be closely related to the reproductive physiological process. Meanwhile, the responsiveness of estrogen treatment in female individual confirmed the above hypothesis of molecular mechanism in regulating sex-specific expression of SOCS1: the typical “bell-shaped” expression kinetics curve was obtained, and the relative expression of SOCS1 reached the peak at 12 h (P < 0.05), suggesting that the expression of SOCS1 may function as an important feedback regulator in the signaling pathway of estrogen signal transduction; while the relative responsiveness of male individual to hormone treatment may indicate the existence of sex-different signal transduction pathway in P. sinensis.
Acknowledgments
This work was supported by University Student Innovation and Entrepreneurship Initiative (S202410549013).
Authors’ Contribution
Writing – original draft: Yulei Zhu; Methodology: Tao Li, Xifan Li and Xiaokai Zheng; Validation: Jingwen Wang; Writing – review & editing: Dan Zeng.
Competing of Interest – COPE
The authors declare that they have no competing interests.
Ethical Conduct Approval – IACUC
Ethical approval for the animal experiments was granted by the Animal Ethics Committee of Hunan University of Arts and Science (HUAS-2024-0032).
Informed Consent Statement
All authors have read and approved the final version of the manuscript for submission.
Data Availability Statement
All are available upon reasonable request.