Introduction
The health and stability of river ecosystems are the cornerstone of aquatic biodiversity. As a key functional group and indicator species in aquatic ecosystems, the changes in the community structure and diversity of fish can effectively reflect the health of the aquatic ecosystem.1 The Fujiang River, an important tributary of the upper Yangtze River, nurtures a rich and unique fish resource, including many endemic species of the upper Yangtze River and nationally protected species, which is of great significance for the biodiversity conservation of the entire Yangtze River basin.2,3 However, from the 20th century to the early 21st century, with the intensification of human activities such as water-related engineering, water pollution, and invasive species, the fish resources of the Fujiang River are facing severe threats.4 Therefore, accurately and efficiently monitoring the dynamics of the fish community in this water body is crucial for formulating scientific conservation strategies and management measures.
Traditional fish survey methods, such as electric fishing, gillnets, and fyke nets, although providing biological information on individual species (e.g., length, weight), have many limitations in practical application.5 These methods are not only time-consuming and labor-intensive but also cause injury or even death to fish, making them unsuitable for routine monitoring of endangered or protected species.6 Furthermore, the capture efficiency of traditional nets is significantly affected by fish size, habits, and the aquatic environment, leading to obvious selective bias and easily missing small, rare, or habitat-specific species, thus underestimating the true species diversity in the region.7
In recent years, environmental DNA (eDNA) metabarcoding technology has emerged as a non-invasive and highly sensitive new method for biological monitoring.8 Organisms release their DNA into the surrounding water through excretion, molting, and reproduction. By collecting water samples, enriching, extracting, and sequencing these eDNA fragments, the species present in the water body can be identified.9 This technology overcomes many of the drawbacks of traditional methods, showing great potential especially in detecting rare, endangered, and cryptic species, and has been widely used in biodiversity assessment and monitoring of rivers, lakes, and oceans.10
This study utilizes eDNA metabarcoding technology to systematically investigate the fish community structure in the Chongqing section of the Fujian River. We collected water samples during the spawning period (June), feeding period (August), and wintering period (November) of the fish, aiming to: (1) Reveal the dynamic changes in the species composition and diversity of the fish community in the Chongqing section of the Fujiang River during different periods, (2) Identify the distribution of key protected species, endemic species of the upper Yangtze River, and invasive alien species in this water body, (3) By comparing with the results of traditional fishing methods in the same period, evaluate the effectiveness and advantages of eDNA technology in monitoring fish diversity in similar river ecosystems. The results of this study will provide a key scientific basis and technical support for the protection of fish resources and the management of the aquatic ecosystem in the Fujiang River and even the upper Yangtze River.
Materials and methods
eDNA sample
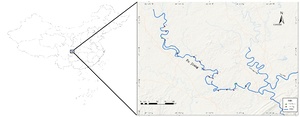
We collected water samples from a total of 14 sampling points along the main channel of the Fujiang River in Chongqing during the spawning period, feeding period, and wintering period of the fish and divided them into 6 river sections. (Figure 1 and Table 1). At each sampling point, a mixed water sample of 2L or 3L were collected from the left, middle and right sections of the river segment, as well as the upper, middle and lower layers of water (if a sampling area has 3 sampling points, then collected 3L of water sample from each sampling point; if a sampling area has 4 or 5 sampling points, 2L of water sample collected from each sampling point). The collected water samples were filtered and enriched on a 0.45 µm mixed cellulose filter membrane using a vacuum pump on site, and the filter membrane was placed in the sample preservation reagent tube.11
Fish sample
In June and October of 2023, operations were conducted using fishing nets in the Chongqing section of the Fujiang River for 7 days, with 10 hours of work each day. We conducted the identification of fish species on the spot, as well as recorded the length and weight of each fish.
DNA extraction and PCR amplification
Microbial DNA was extracted from filter membrane samples using the E.Z.N.A.® Water(DNA Kit (Omega Bio-tek, Norcross, GA, U.S.) according to manufacturer’s protocols. The function gene were amplified by PCR (95 °C for 2 min, followed by 25 cycles at 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s and a final extension at 72 °C for 5 min) using primers freshwater fish 12S Tele02_F 5′-AAACTCGTGCCAGCCACC-3′,Tele02_R 5′-GGGTATCTAATCCCAG TTTG-3′, and COI MlCOIintF 5’-GGW ACWGGW TGAACWGTWTAYCCYCC-3’,JghHCO2198 5’-TAIACYTCIGGRTGICCR AARAAYCA-3’ where barcode is an eight-base sequence unique to each sample. PCR reactions were performed in triplicate 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase, and 10 ng of template DNA. Amplicons were extracted from 2% agarose gels and purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, U.S.) according to the manufacturer’s instructions.12
Library Construction and Sequencing
Purified PCR products were quantified by Qubit®3.0 (Life Invitrogen), and every twenty-four amplicons whose barcodes were different were mixed equally.The DNA was fragmented using Ultrasonic instrument.13 A DNA library was obtained by unwinding the DNA duplex product from PCR amplification and disrupting the uncircularized DNA molecules. The libraries were finally sequenced using next generation sequencing platform (MGI-G99 or Illumina Miseq) (Shanghai BIOZERON Biotech. Co., Ltd) with PE300 mode according to the standard protocols.14
Processing of sequencing data
Raw fastq files were first demultiplexed using Trimmomatic5 (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic) and in-house perl scripts according to the barcode sequences information for each sample with the following criteria: (i) The 300bp reads were truncated at any site receiving an average quality score <20 over a 10 bp sliding window, discarding the truncated reads that were shorter than 50bp. (ii) exact barcode matching, 2 nucleotide mismatch in primer matching, reads containing ambiguous characters were removed. (iii) only sequences that overlap longer than 10 bp were assembled according to their overlap sequence. Reads which could not be assembled were discarded.15,16
Results
Fish composition based on eDNA
The composition of fish during the spawning period
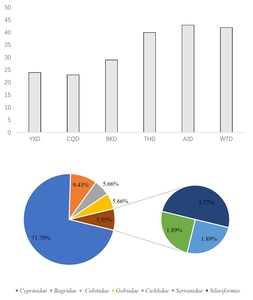
In June, we used eDNA technology to detected 72 species of fish in six sections of the Fujiang River. They belong to 11 families and 52 genuses. It includes two species, Leptobotia elongate and Procypris rabaudi, which are both classified as second-level protected animals in the country. They also include five types of municipal key protected fish species: Liobagrus marginatus, Xenocypris fangitchang, Ochetobius elongatus, Leptobotia microphthalma, and Leptobotia taeniops. There are 15 species of fish unique to the upper reaches of the Yangtze River Leptobotia elongata, Procypris rabaudi, Xenocypris fangitchang, Leptobotia microphthalma, Hemiculterella sauvagei, Paracobitis potanini, Ancherythroculter kurematsui, Ancherythroculter nigrocauda, Megalobrama Pellegrini, Botia reevesae, Liobagrus marginatoides, Sinogastromyzon sichangensis, Rhinogobio cylindricus, Acrossocheilus monticolus, Hemiculter tchangi. There are 5 invasive species: Mrigalcarp, Pseudohemiculter dispar, Coptodon zillii, Gambusia affinis, Silver prussian carp. The most common family was Cyprinidae with 47 species, accounting for 65.28%; the second family was Bagridae with 7 species, accounting for 9.72%; the proportion of fish species that are herbivorous was 29.60% (Figure 2).
The composition of fish during the Feeding migration period
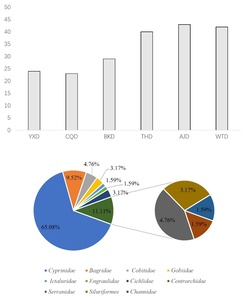
In August, using eDNA technology, we identified a total of 53 fish species, belonging to 7 families and 41 genera. They include one species of nationally second-level protected fish, Procypris rabaudi. They also include two types of municipal key protected fish species: Xenocypris fangitchang, Ochetobius elongatus,There are 7 species of fish unique to the upper reaches of the Yangtze River Procypris rabaudi, Xenocypris fangitchang, Hemiculterella sauvagei, Ancherythroculter kurematsui, Ancherythroculter nigrocauda, Bangana rendahli, Hemiculter tchangi. There are 4 invasive species: Pseudohemiculter dispar, Nile tilapia, Megalobrama terminalis, Silver crussian carp. The most family was Cyprinidae with 38 species, accounting for 71.70%; the second family was Bagridae with 5 species, accounting for 9.43% (Figure 3).
The composition of fish during the overwintering period
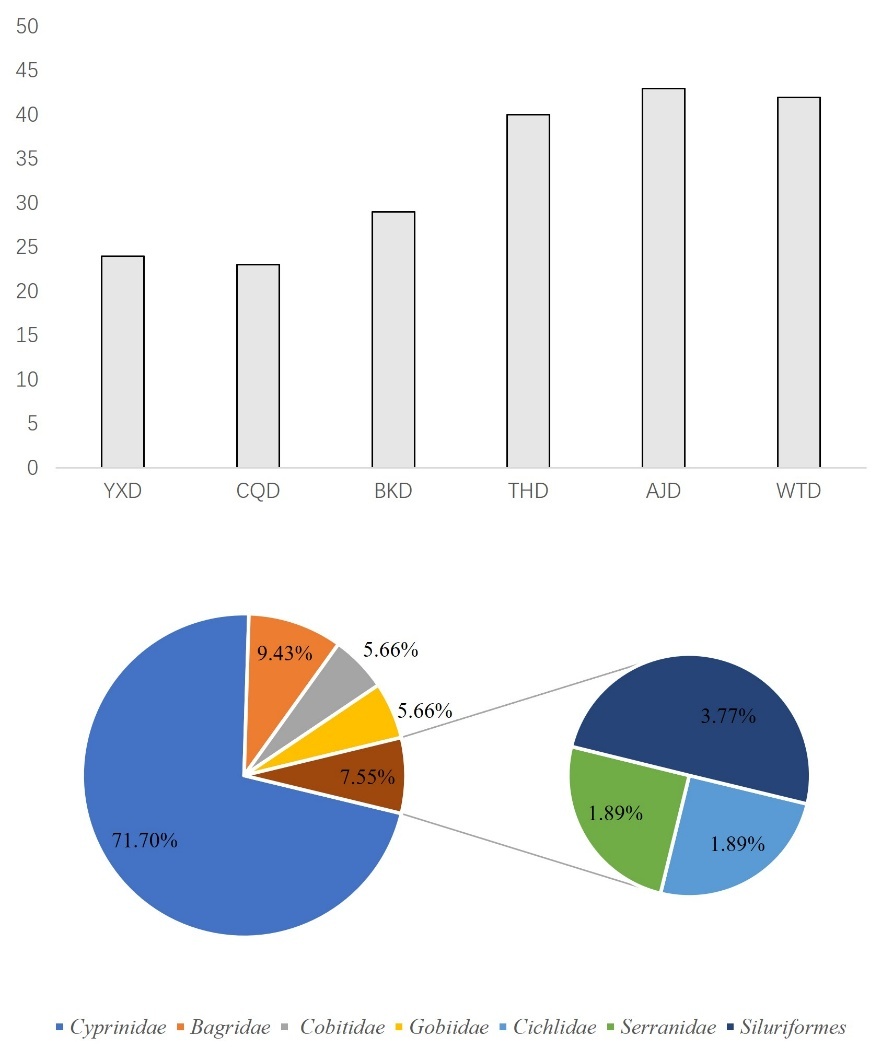
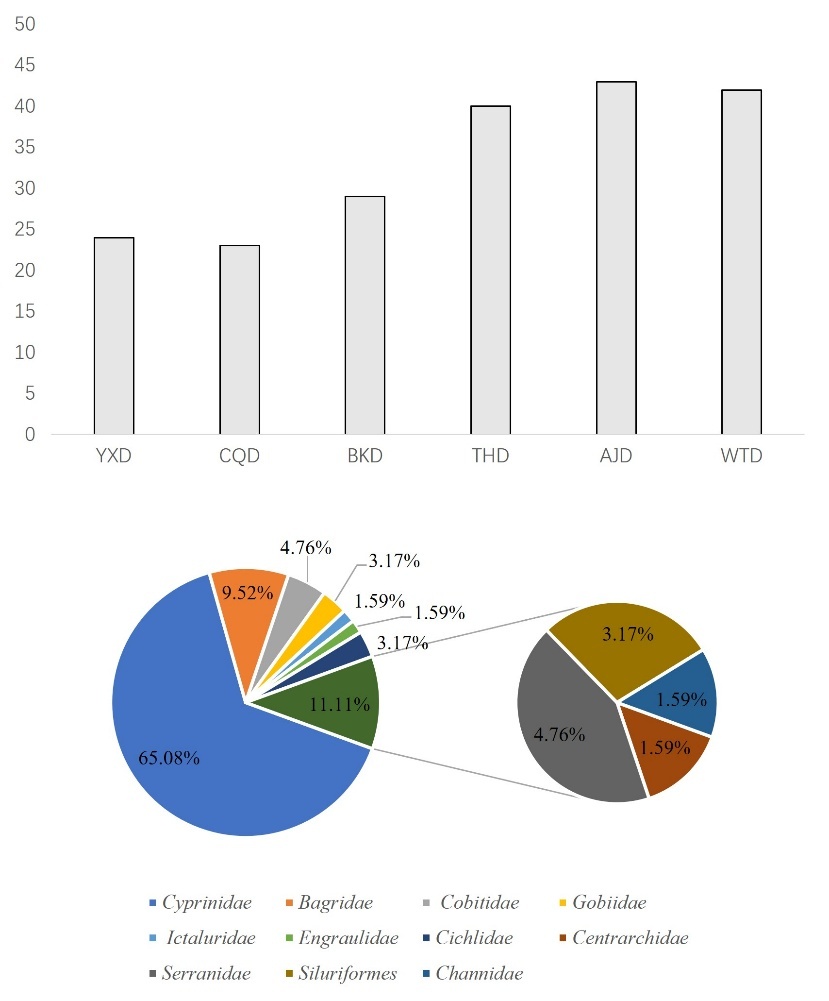
In November, using the eDNA technology, we identified a total of 63 species of fish belonging to 11 families and 55 genera. Among them, there are 63 species of indigenous fish, including 1 species that is a national second-level key protected fish. They include one species of nationally second-level protected fish, Procypris rabaudi. They also include two types of municipal key protected fish species: Xenocypris fangitchang, Ochetobius elongatus. There are 5 species of fish unique to the upper reaches of the Yangtze River Procypris rabaudi, Xenocypris fangitchang, Ancherythroculter nigrocauda, Acrossocheilus monticolus, Hemiculter tchangi. There are 8 invasive species: Ictalurus punctatus, Micropterus salmoides, Cirrhinus molitorella, Pseudohemiculter dispar Coptodon zillii, Nile tilapia, Megalobrama terminalis, Silver prussian carp. The most family was Cyprinidae with 41 species, accounting for 65.08%; the second family was Bagridae with 6 species, accounting for 9.52% (Figure 4).
The composition of fish species based on traditional fishing methods
The composition of fish during the spawning period
In June, we caught a total of 40 species of fish in the Fujiang River. Among them, all were native species, and there was 1 species of nationally second-class protected fish Procypris rabaudi. There are 7 species of fish unique to the upper reaches of the Yangtze River: Ancherythroculter kurematsui, Ancherythroculter nigrocauda, Megalobrama pellegrini, Hemiculter tchangi, Procypris rabaudi, Gnathopogon herzensteini. The most family was Cyprinidae with 30 species, accounting for 75.00%; the second family was Bagridae with 6 species, accounting for 15.00%. The proportion of fish species with a carnivorous diet accounted for 37.50%, and the Shannon-Wiener index was 3.05.
The composition of fish during the Feeding migration period
In October, we caught a total of 37 species of fish in the Fujiang River. Among them, 36 were native species, and 1 was an introduced species. there was 1 species of nationally second-class protected fish Procypris rabaudi. And there are 6 species of fish unique to the upper reaches of the Yangtze River: Ancherythroculter kurematsui, Ancherythroculter, Nigrocauda, Megalobrama, Pellegrini,Hemiculter tchangi, Procypris rabaudi, Xenocypris yunnanensis. The most common family was Cyprinidae with 26 species, accounting for 70.27%; the second family was Bagridae with 5 species, accounting for 13.51%. The proportion of fish species with a carnivorous diet accounted for 40.50%, and the Shannon-Wiener index was 2.89.
Analysis of Fish Diversity in the Chongqing of the Fujiang River Based on eDNA
The alpha diversity indices we use are calculated based on the OTU (operational taxonomic unit) table obtained from eDNA research.17,18 These indices measure the diversity of the community from different perspectives: The results of α diversity analysis showed that among the 14 sampling points, the Chao1 index ranges for June, August and November were 12-45, 21-41 and 18-33, respectively. The Shannon-Wiener index ranges for June, August and November were 0.61-2.71, 0.45-1.98 and 0.53-2.59, respectively. The Simpson index ranges for June, August and November were 0.25-0.91, 0.19-0.82 and 0.19-0.91, respectively (Table 2).
The α diversity indices of fish species at each section show certain differences. The α diversity indices at the same section also vary at different times. Based on the results from different sampling points across the three periods, FJ-10 exhibits a relatively high Chao1 index. FJ-7 and FJ-10 have relatively high Shannon-Wiener indices, while FJ-7, FJ-9, and FJ-10 have relatively high Simpson indices. In the Fujiang river, considering the three periods of the entire river, August has a relatively high Chao1 index, while November has relatively high Shannon and Simpson indices.
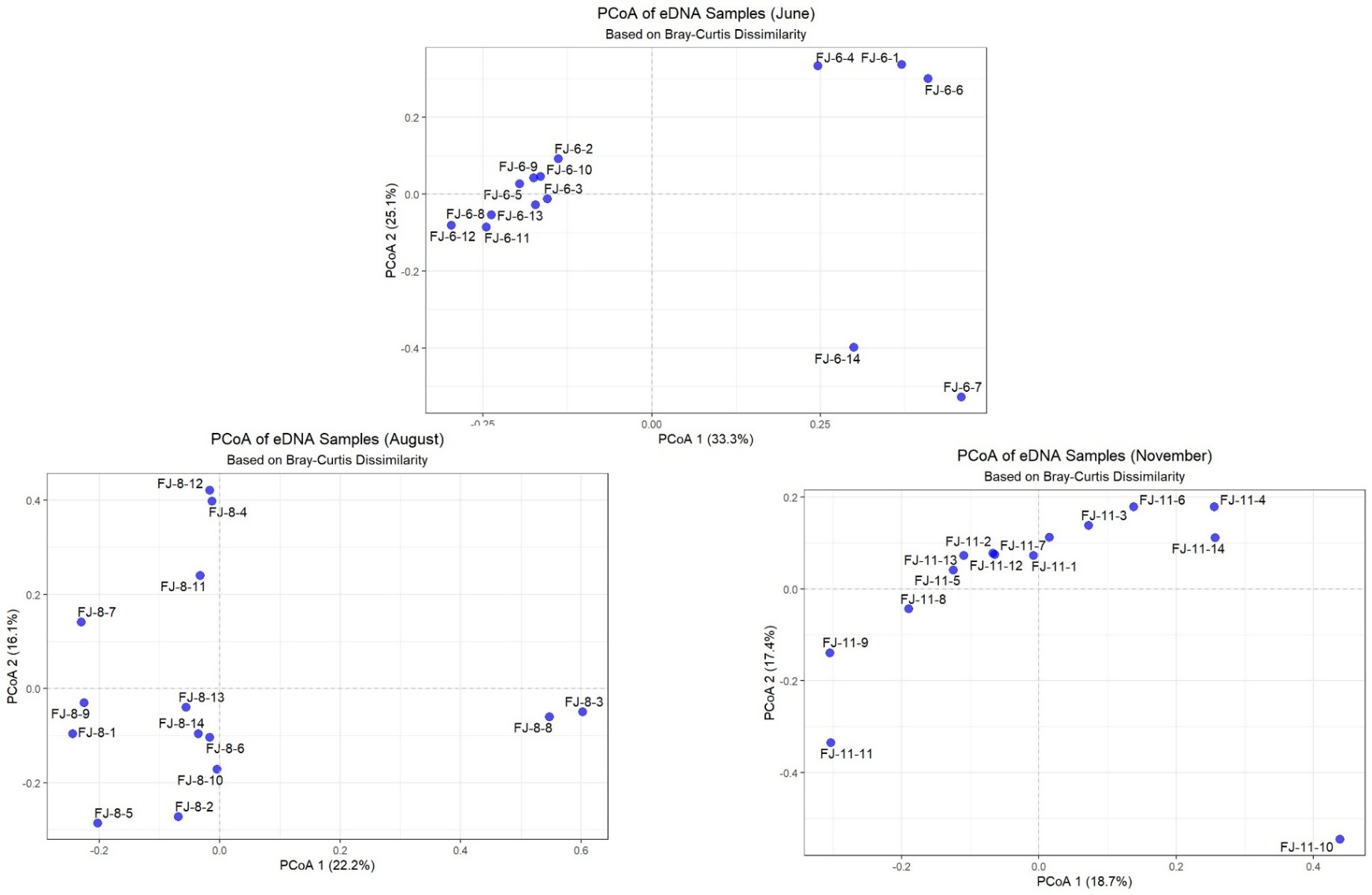
Principal coordinate analysis (PCoA) based on Bray-Curtis distance revealed that the fish community structure in the Chongqing section of the Fujiang River exhibited a strong seasonal dependence: the first two axes accounted for up to 58.4% of the variance in June (the spawning period), and the samples showed a distribution pattern of three isolated islands. As the breeding period comes to an end and the period from August to November begins, the explanation rate drops to around 36%, and the community distribution gradually shifts from a “severely fragmented” state to a “continuous gradient” along the axial direction (Figure 5).
Discussion
This study is the first to utilize eDNA metabarcoding technology to systematically investigate the fish community composition in the Chongqing section of the Fujiang River in different seasons and compare it with traditional fishing methods. The results demonstrate that eDNA technology offers significant advantages in species detection ability, monitoring efficiency, and the detection of both protected and invasive species.
The High Efficiency and Sensitivity of eDNA Technology in Fish Diversity Monitoring
Compared with traditional fishing methods, eDNA technology has shown a higher rate of species detection. Throughout the study period, eDNA technology detected far more fish species than traditional fishing methods (for example, in June alone, eDNA detected 72 species, while traditional methods only caught 40 and 37 species in August and October, respectively). This difference highlights the limitations of traditional methods, specifically the selective bias of nets for fish size, life habits (such as benthic or burrowing), and swimming ability. By directly detecting species signals in the water, eDNA technology can more comprehensively cover all species in the habitat, especially small fish, rare populations, or fry that are difficult to capture by traditional methods.19
Of particular importance, eDNA technology successfully detected a variety of protected species, such as the national second-class protected animal Leptobotia elongata and Procypris rabaudi, as well as a variety of municipal key protected fish species. Among them, Leptobotia elongata did not appear in the catch of traditional fishing methods, which indicates that eDNA technology has irreplaceable value for monitoring rare and endangered species with cryptic behavior and low population density, and can provide earlier and more accurate early warning information for species protection.20
In addition, eDNA technology has also effectively identified a variety of invasive alien species, such as Coptodon zillii, Gambusia affinis, and Nile tilapia. The invasion of alien species is one of the important factors leading to the decline of native fish populations. eDNA can facilitate rapid screening and early detection of invasive species, which is crucial for preventing their further spread and informing the development of effective control measures.
Seasonal Dynamics of Fish Community Composition
The eDNA results of this study revealed obvious seasonal changes in the fish community of the Fujiang River. The species richness peaked in June (spawning period) (72 species), dropped to its lowest in August (feeding period) (53 species), and recovered somewhat in November (wintering period) (63 species).
The high species diversity in June may be attributed to the increased activity, schooling, and migration behavior of fish during the spawning period, which increases the amount and diffusion range of DNA released into the environment. In addition, suitable water temperature may also promote the release of DNA.21 The decrease in the number of species in August may be related to a variety of factors: first, the hydrological conditions of high flow velocity and large flow during the summer flood period may accelerate the dilution and degradation of eDNA, reducing the detection rate; second, some fish may enter tributaries or specific habitats for feeding, resulting in an uneven distribution in the main stream. The recovery of the number of species in November may be due to the fact that fish activity decreases and tends to congregate in deep and slow-flowing river sections during the wintering period, which makes eDNA relatively enriched in local areas, thereby increasing the probability of being detected.
The seasonal changes in the α diversity index also confirm this point. The Chao1 index was relatively high in August, while the Shannon and Simpson indices were relatively high in November. This indicates that although the number of species actually detected in August was small, there may be more rare species signals in the water that have not been fully detected (high Chao1); while the fish community structure in November showed higher evenness (high Shannon and Simpson), which may be related to the ecological habit of different species mixing and gathering during the wintering period.17
Conclusion
This study systematically evaluated the application of eDNA metabarcoding technology in monitoring fish diversity in the Chongqing section of the Fujiang River. The main conclusions are as follows: Compared with traditional fishing methods, eDNA technology is a more sensitive, efficient, and non-invasive monitoring tool. It significantly improves the detection rate of species, especially showing great advantages in detecting rare, endangered, and invasive alien species. The fish community structure in the Chongqing section of the Fujiang River shows obvious seasonal dynamics. Species richness is highest during the spawning period (June), lowest during the feeding period (August), and recovers during the wintering period (November), which is closely related to the life activities of fish and changes in the hydrological environment. This study successfully utilized eDNA technology to identify national key protected fish species, such as Leptobotia elongata and Procypris rabaudi, as well as a variety of endemic species from the upper Yangtze River and invasive alien species, providing key information for the targeted protection and management of these species. In summary, eDNA technology provides a powerful technical means for the rapid and comprehensive assessment of river fish biodiversity. It is recommended that this technology be integrated into the routine system of aquatic biological monitoring in the Fujiang River and the entire Yangtze River basin in the future to support the protection and sustainable development of the aquatic ecosystem in this region.
Acknowledgments
The authors extend their sincere gratitude for the technical assistance rendered by the Chongqing Fisheries Science Research Institute
Authors’ Contribution
Conceptualization: Qianqian Ma (Equal), Yan Dan (Equal). Software: Qianqian Ma (Equal), Yong Xie (Equal). Writing – original draft: Qianqian Ma (Lead). Writing – review & editing: Qianqian Ma (Equal), Yan Dan (Equal), Chuang Zhang (Equal), Yan Li (Lead). Data curation: Yong Xie (Lead). Formal Analysis: Yong Xie (Equal). Resources: Yan Li (Lead).
Competing of Interest – COPE
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Ethical Conduct Approval – IACUC
No approval from research ethics committees was required for this study, as the fish samples were collected from commercial catches made by the fishers.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.