Introduction
Streptococcus iniae is one of the most significant bacterial pathogens in aquaculture, causing widespread disease in marine and freshwater fish species. This Gram-positive bacterium has been linked to severe outbreaks, particularly in intensively farmed species, resulting in substantial economic losses. The disease is characterized by septicemia, neurological signs, and skin lesions, and often results in high mortality. In marine fish farming, S. iniae is a well-documented cause of streptococcosis, with outbreaks reported in multiple species, including Asian seabass (Lates calcarifer), snapper (Lutjanus spp.), and tilapia (Oreochromis niloticus). Mortality during outbreaks often ranges from 30% to 50%, with some cases exceeding 70%, depending on environmental factors and management practices. A study on Asian seabass farms in Southeast Asia reported losses with annual losses exceeding $10 million due to S. iniae infections.1–3
Asian seabass is highly susceptible to S. iniae, particularly in intensive farming systems. Stressors such as high stocking density, fluctuating temperatures, and poor water quality exacerbate the risk of infection. For instance, farms in Australia have documented mortality rates of 35-50% during severe outbreaks, resulting in production losses of several hundred tons per year.4 In Vietnam, S. iniae was reported in cage-raised Asian seabass in Khanh Hoa province,5 and Thua Thien Hue province,6 causing great economic losses to farmers. Histopathological findings in infected Asian seabass include granulomatous inflammation in the kidney and spleen, with bacterial colonies present in multiple organs. The disease often progresses rapidly, with signs such as erratic swimming, exophthalmia, and hemorrhages appearing within days of exposure.
Globally, the cost of managing S. iniae in aquaculture, including treatment, preventive measures, and production losses, is estimated to exceed $100 million annually. Asian seabass farming operations in Southeast Asia and Australia are particularly affected, where frequent outbreaks of disease disrupt supply chains and reduce profitability.7
Vaccination is one of the most effective strategies to control Streptococcus iniae infections in aquaculture. Over the years, various vaccine types, including inactivated, subunit, and recombinant vaccines, have been developed and tested in marine fish, with varying degrees of success. Inactivated vaccines are the most commonly used in aquaculture due to their safety and ease of production, resulting in a 60-80% reduction in mortality in field trials.2,8
However, understanding the biochemical, genetic, virulence, and immunogenic properties of S. iniae is crucial for selecting suitable strains for inactivated vaccine production. Sampling of diseased fish from farms in Vietnam, isolation, identification of bacteria, and assessment of pathogenicity in healthy fish were performed (data not shown). This study was conducted to confirm the virulence and immunogenicity properties of strain S. iniae SiTH1, to select a highly relevant strain as a candidate for inactivated vaccine production.
Materials and Methods
Bacterium
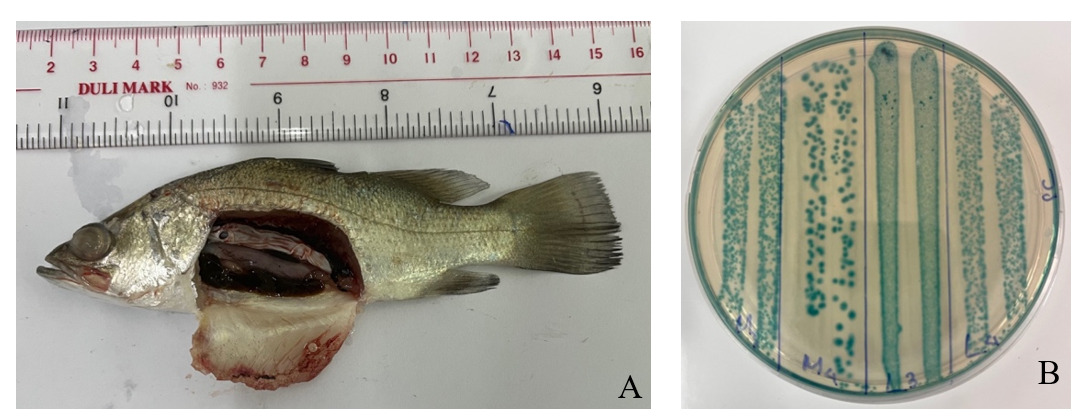
S.iniae SiTH1 was isolated from Asian seabass cultured cages in Vietnam. Infected fish showed clinical signs such as lethargy, disoriented swimming, skin hemorrhages and pop-eyes. The S. iniae strain SiTH1 was identified by biochemical characteristics using the API 20 strep kit (BioMerieux, France) and 16S rRNA gene sequence analysis (GenBank accession no. OR192844.1).
S. iniae SiTH1 strain was cultured in BHI broth (Merck) supplemented with 1.5% NaCl and incubated at 30 °C. After 36 hours of incubation, the harvested bacterium suspension was supplemented with 20% glycerol (v/v), divided into cryotubes, and stored at -80 °C for further studies.
Survey species-specific identifying genes of S. iniae
S. iniae SiTH1-specific genes were identified by primer pairs including ITS rDNA9 and lactate oxidase (lctO) genes.10 Accordingly, the strain was cultured in BHI broth medium for 36h, centrifuged at 1,000 xg, 4 °C for 10 minutes. DNA was extracted from precipitated bacteria using the GeneJET Genomic DNA Purification kit (Thermo, USA), following the manufacturer’s instructions. The DNA product was used for PCR reactions, shown in Table 1. The PCR products of ITS rDNA and lctO genes were visualized by electrophoresis on a 1.5% agarose gel stained with ethidium bromide using the GelDoc system (Carestream Health, Inc. – Kodak/USA).
Survey the virulence genes of S. iniae SiTH1
The S. iniae SiTH1 was investigated for the presence of seven virulence genes of S. iniae simA, pdi, pgm, cpsD, scpI, sagA, and cpsA in a multiplex PCR, following the method of Deng et al.11 and Park et al.12 The strain was cultured, and DNA was extracted as described above. The DNA product was used for the PCR reactions to survey virulence genes, as shown in Table 2, and PCR products were visualized by electrophoresis as described above.
Experimental challenge
Healthy seabass fingerlings weighing 10-15 g, 8-10 cm in length, were acclimatized for 14 days in a composite tank containing seawater with a salinity of 32-33 ‰, temperature of 27-28 °C, continuous aeration, and fed 3% BW once a day by commercial pellet feed (C.P. Vietnam Corporation). After the acclimatization period, three fish were randomly selected for bacterial isolation to confirm the absence of Streptococcus infection.
A colony of S. iniae SiTH1 was cultured in BHI broth at 30°C for 36 hours. Then, the bacterial suspension was centrifuged at 1,000 xg, 4 °C for 10 minutes, and the precipitated bacteria were resuspended in sterile PBS, adjusted spectrophotometrically equivalent to 108 CFU/mL (OD600 = 1.0, approximately, based on growth curve data). Then, the bacterial suspension was diluted in PBS to the challenge concentrations of 104, 105, 106, and 107 CFU/fish for identification of the lethal challenge dose of 50% (LD50). Accordingly, each injection concentration was repeated in triplicate, and each tank contained 10 fish. Each fish was injected intraperitoneally (i.p) with bacterial suspension as challenge concentrations (0.1 mL). The fish control group was injected i.p with sterile PBS instead of bacteria. Each group was reared in a separate 150 L composite tank installed with a water circulation system. Clinical signs and mortality of the fish were monitored for 14 days. During the experimental period, moribund and dead fish were rapidly removed from the tank for bacterial isolation. The lethal challenge dose of 50% (LD50) was calculated following Reed and Muench.13
Vaccine and immunization of fish
S. iniae SiTH1 was cultured in BHI broth at 30°C for 36 hours. Then, the bacterial suspension was inactivated by supplementing 1% formalin (FKC - Formalin killed cell) over night at 4 oC, sampled to culture on BHI broth and BHI agar at 30°C for 48 hours to confirm absolutely inactivated bacteria, then centrifuged at 1,000 xg, 4oC for 10 minutes, the precipitated bacteria was suspended and diluted in sterile PBS for the experimental antigen concentrations of 109, 1010 and 1011 CFU FKC/mL, corresponding to treatments A9, A10, A11, respectively.
The fish immunization experiment was conducted using seabass fingerlings weighing 10-15 g and 8-10 cm in length, divided into nine composite tanks, with 35 fish/tank, corresponding to the three above treatments (A9, A10, A11), in triplicate. Fish were injected with FKC of different antigen concentrations by i.p injection of 0.1 mL/fish. The non-vaccinated group (Non-A) was similarly treated but received injections of PBS. All fish were fed 3% BW once a day.
Evaluation of immune gene expression
At 24h and 21 days post-immunization, 5 fish/tank were anaesthetised and sacrificed for kidney samples to evaluate expression levels of some immune-related genes. Briefly, mRNA was extracted from the kidney samples using Total RNA kit I (Promega) according to the manufacturer’s instructions and quantified using a Nanodrop spectrophotometer (Thermo Scientific). For cDNA synthesis, 5 µg of the sample was used as the template (Script cDNA synthesis kit - Biorad), following the manufacturer’s instructions, and then diluted and stored at -20 °C. Quantification of mRNA by RT-qPCR using QuantiTect® SYBR Green PCR Master Mix (Qiagen) amplified five immune-related genes in sea bass, including interleukin 1β (IL-1β); Interleukin 6 (IL-6); Interleukin 10 (IL-10), Mx protein (Mx); and Tumor necrosis factor α (TNF-α). The primer sequences, components, and thermal cycling of the reaction were performed according to the instructions of Aamri et al.14 Expression analysis as relative amounts of the target gene using β-actin as the internal reference gene, the content in each sample was calculated using the comparative Ct method (2-ΔΔ*Ct*).15
Serum collection and Enzyme-linked immunosorbent assay (ELISA)
At 21 days post-immunization, 5 fish/tank were anaesthetised and blood collected from the caudal vein for serum separation. The IgM antibody in serum was analyzed by the indirect enzyme-linked immunosorbent assay (ELISA).16 To evaluate the appropriate dilution for the ELISA assay, serum samples were pooled from three fish for each treatment, and 2-fold serially diluted solutions were used. The suitable dilution for ELISA analysis for serum was found to be 1:3200. To analyze humoral antibody in serum samples, inactivated S. iniae strain SiTH1 (40 µg/mL) was added to a 96-well plate (100 µL/well), and incubated overnight at 4°C. Then, a blocking solution, PBS-T containing 3% Skimmed milk, was supplied for 1 hour at room temperature. Seabass serum (diluted 1:3200) was added (50 µL/well, repeated three times/sample), and the plate was incubated for 2 hours. This was followed by Mouse anti-Asian seabass (Lates calcarifer) IgM monoclonal antibody (F02-Aquatic diagnostics Ltd, diluted 1:40 in PBS-T), incubated for 2 hours, then Polyclonal Rabbit anti-mouse Ig-HRP polyclonal antibody (Dako, diluted 1:2000 in PBS-T), incubated for 1 hour. Between steps, wells were washed 3 times with PBS-T. The enzymatic reaction was activated with O-Phenylenediamine substrate in phosphate-citrate buffer (50 µL/well) and stopped after 10 min with 50 µL of 2.0 M H₂SO₄. Optical density was measured at 492 nm using an iMark reader (Bio-Rad).
Relative percent survival (RPS)
At 21 days post vaccination, 25 fish/tank vaccinated with antigen concentrations (109; 1010; 1011 CFU FKC/mL, corresponding to treatment A9, A10, A11, respectively) and a non-vaccinated group (Non-A) were challenged by i.p. injection of 0.1 mL/fish of wild-type S. iniae SiTH1 at a concentration of 108 CFU/mL. At the same time, a control group (C-PBS) was similarly arranged and injected with PBS. Mortalities were recorded daily for 14 days, and RPS was calculated using the following function.
RPS = (1- (% mortality of the vaccinated group/% mortality of the non-vaccinated group)) * 100.
Data analysis
The data were statistically analyzed with the statistical analysis program SPSS, version 20.0. All data are represented as Mean ± SE. ANOVA one-way (analysis of variance) and Tukey HSD post hoc tests were used to determine significant differences between groups in experiments, at P<0.05.
Results
Species-specific genes of S. iniae
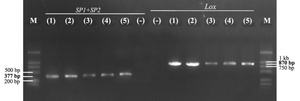
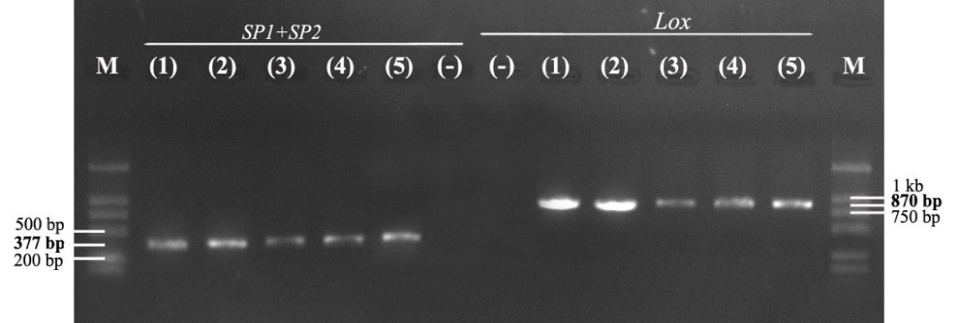
The results of electrophoresis of PCR products with two specific primer pairs, SP1/SP2 and LOXF/LOXR, showed that the strain SiTH1 had two target gene segments, ITS rDNA and lctO, with PCR product sizes of about 377 bp and 870 bp, respectively (Fig. 1). Thus, the PCR method with primer pairs specific to the target gene segments ITS rDNA and lctO, two highly conserved genes, identified the species quickly and effectively.9,10
Virulence genes
S. iniae bacteria contain virulence factors regulated by genes that play different roles in the process of infection and disease in fish. The results showed that strain S. iniae SiTH1 contains six out of seven tested virulence genes, including simA, scpI, pdi, pgm, cpsD, and sagA. The identified PCR products are shown in Figure 2.
Experimental challenge
The results of fish challenge tests showed rapid mortality, with the highest mortality occurring at high infectious doses. A gradual stable survival thereafter from about 10 days post-infection (dpi), indicating the acute virulence of the bacterial strain to fish (Fig. 3). The 50% lethal dose (LD50) was calculated as 104.4 CFU/fish. Dead fish of challenged test were immediately sampled for bacterial analyses, and all showed characteristic colonies of S. iniae bacteria with up to 104-106 CFU/g of kidney, spleen, brain, and eyes (Fig. 4).
Immune gene expression in seabass after immunization with FKC
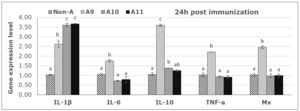
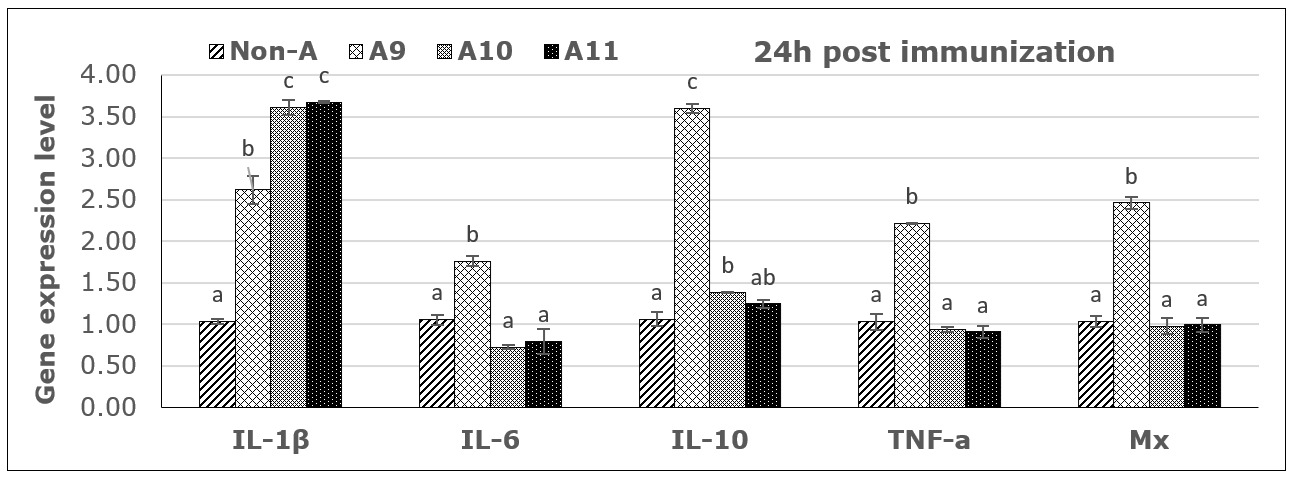
From PCR products and analysis of the expression levels of five immune genes by RT-PCR, all immune genes were activated 24 hours post-vaccination, as shown in Figure 5. However, the expression trends were different between immune genes and depended on the antigen concentrations. Specifically, the immune gene IL-1β was activated after 24 hours at all three antigen concentrations, with the concentrations of 1010 and 1011 CFU FKC/mL providing a 3.5-fold upregulation compared to the control, followed by the concentration of 109 CFU/mL, which was 2.5 times higher than the control. For the immune genes IL-6, TNF-α, and Mx, expression was only activated after 24 h at a concentration of 109 CFU FKC /mL. At the same time, there was almost no activation or inhibition at the two higher concentrations. For the immune gene IL-10, the results showed that there was a strong activation after 24h at concentrations of 109 and a mild activation in higher concentrations (Fig. 5).
Results of immune gene expression analysis at 21 days post-vaccination showed that only the IL-1β and TNF-α genes were detected, while the Mx, IL-6, and IL-10 genes did not show any signals in the analyzed groups (no data displayed). Accordingly, the vaccine with concentrations of 109 and 1010 CFU FKC/mL still yielded the best results in stimulating immune gene expression.
Humoral antibody response
Serum antibody levels were analysed at 21 days post-vaccination. The antibody levels in fish sera tended to increase gradually with antigen concentration, being highest in the group injected with a vaccine concentration of 1011 CFU FKC /mL, reaching 0.36 (OD492nm) (Table 3).
Disease resistance
Fish in the vaccinated groups maintained a stable high survival after 7 days; fish survival improved with increasing immunization doses of 109, 1010, and 1011 CFU FKC/mL, where survival rates were 70.7%, 81.3%, and 82.7%, respectively. In the negative control group, which was injected with PBS, all fish survived (100%) throughout the entire monitoring period. Meanwhile, fish in the non-vaccinated group, injected with wild-type bacteria SiTH1, began to die 24 hours post-challenge and reached 80% mortality at 8 days post-injection (dpi). From the above results, the relative percent survival (RPS) in fish after 21 days of immunization at vaccine concentrations of 109 CFU FKC/mL (A9), 1010 CFU FKC/mL (A10), and 1011 CFU FKC /mL (A11) reached 63.3%, 76.7%, and 78.3%, respectively (Table 4).
Discussion
S. iniae have been reported to be the causative agent of severe disease in many marine fish species, often externally identified in infected sea bass with a typical sign of pop and opacity eyes. Studies on isolation and identification of this bacterial species are based on biochemical characteristics that can easily be confused, so determining genetic characteristics based on specific primer pairs SP1/SP2 and LOXF/LOXR to detect two target gene segments, ITS rDNA and lctO, respectively, helps to identify S. iniae accurately and quickly. The strain S. inae SiTH1 was found to be positive using these two specific primer pairs, demonstrating its high effectiveness and reliability in the rapid identification of S. iniae bacteria. These results are in accordance with several previous studies that presented PCR results using the two specific primer pairs mentioned above, such as Creeper & Buller,17 Suanyuk et al.,18 and Zhou et al.9
These studies also confirmed that the isolated strains contained the virulence genes, often found in highly virulent strains, when testing for infectivity by experimentally infecting healthy fish. Many studies have shown that S. iniae contains virulence factors regulated by genes with functions that perform different roles in the process of infecting and causing disease in fish. Strains containing a greater number of toxin genes, associated with the ability to invade fish and cause disease, have 7 toxin-encoding genes (simA, pdi, pgm, sagA, cpsA, cpsD, and scpI). They contribute to different functions: simA is involved in adhesion, penetration, and growth; pdi deals with protection against lysozyme degradation; pgm contains functions against enzymes and antibacterial peptides; sagA causes damage to host cells; cpsA, cpsD, and scpI are extracellular polysaccharides involved in escaping phagocytosis. A strain containing a variety of toxin genes is always a good candidate for screening to select a representative and relevant strain for testing of vaccine antigen candidates. The strain S. iniae SiTH1 in this study contains 6 of 7 toxin-encoding genes, including simA, scpI, pdi, pgm, cpsD, and sagA. Similarly, these 6 toxin-encoding genes have been identified in 2 strains of S. iniae, Ab130920 and Ab131025, isolated from Siberian sturgeon (Acipenser baerii) in China,11 and S. iniae K288 strain isolated from Nile tilapia (Oreochromis niloticus).19 The virulence of the S. iniae SiTH1 strain was also confirmed through the experimental challenge of healthy fish. The fish began to weaken and die 24 hours post challenge and reached a mortality rate of over 50% at 7 dpi with a lethal dose of LD50 of 104.4 CFU/fish. Such mortality and LD50 is considered to show highly toxic effects to marine fish. Some studies have also reported similar results on the LD50 value of S. iniae in seabass distributed in Vietnam. A survey of seabass in the south central region of Vietnam (Nha Trang - Khanh Hoa) showed LD50 value of from 104,8 to 105,8 CFU/fish,5 and from seabass distributed in the north central region of Vietnam (Hue), the LD50 was 105.28 CFU/fish.6
In the present study, seabass immunized with inactivated bacteria at different concentrations also showed the ability to stimulate immune-related genes. Interleukin-1β was clearly activated after 24 hours at different antigen concentrations, with high expression levels. In contrast, immune genes IL-6, TNF-α, Mx, and IL-10 were only strongly activated at lower concentrations. The mild activation of the high antigen dose may indicate a dose-response in this species due to tolerance or the induction of inhibitory regulations to high antigen concentrations, as is known for animals and humans. The specific mechanisms or cells involved in fish are not well understood, but they may involve inflammatory reactions and T-cell regulatory mechanisms to inhibit unfavorable immune responses. However, the ability to stimulate humoral antibody production in fish and the high protective effect were achieved at antigen concentrations of 1010 and 1011 CFU/mL. A similar study in rainbow trout tested an inactivated vaccine based on the S. iniae strain Dan-1 prepared at three antigen concentrations of 3x109, 3x1010, and 3x1011 CFU/mL. After 1 month post vaccination, a bacterial challenged experiment showed that the survival rate of fish receiving vaccine doses of 1010 and 1011 CFU/mL both reached 90% survival, while the vaccine dose of 109 only reached 50% survival; the non-vaccinated control group had 20% survival.20 Another study demonstrated the effect of different bacterial concentrations on tilapia immunity and optimal vaccine concentration to induce immunity in Nile tilapia. The experiment was performed at 102, 104, 106, 108, and 1010 CFU/fish of S. agalactiae compared with the control (PBS) through intraperitoneal injection for 72 h. The statistic revealed a significant difference (p<0.05) in the 108 and 1010 CFU/fish injections with high survival rates (62.22% and 53.33%, respectively) as well as immunoglobulin gene expression and antibody titer were highly represented in the 1010 CFU/fish injection.21 A previous study also reported the efficacy of inactivated vaccines against S. iniae in seabass (Lates calcarifer), fish were vaccinated with i.p 1012 CFU/fish, revealed that no mortality at all time points (4, 8, 12, 20, and 28 weeks post-vaccination and challenge), while the control fish presented 10–43.33% mortality. And antibody levels in vaccinated fish increased significantly and innate immune genes, including MHC I, MHC II, IL-1β, IL-4/13B, and IL-10, were significantly upregulated at 12 h.8 The protective effect of the vaccine tested in this experiment is comparably practical to fish vaccines used for several fish species when tested under experimental conditions and for commercial fish vaccines. The present initial experimental study aimed to identify a suitable bacterial strain, isolated from infected fish in local farms, that could be used to elicit immune responses and induce protective immunity in seabass. The results obtained regarding antibody response and protection short time after vaccination, provide a solid foundation for further testing of the vaccine candidate, which includes adjuvants and field testing where long-term immunity can be shown.
In conclusion, Streptococcus iniae SiTH1 has all the characteristics, including being a pathogenic strain with disease signs similar to those found in natural disease outbreaks. The strain contains multiple toxin genes and can elicit strong immune responses, resulting in increased humoral antibody levels and protective immunity in fish. The Streptococcus iniae SiTH1 strain should be considered a good candidate for inactivated vaccine production against streptococcosis in seabass in Vietnam.
Acknowledgments
This research was supported by a grant from the Ministry of Agriculture and Environment for the Research Institute for Aquaculture No.3, Vietnam, grant number: 45/2022/HĐ-KHCN-TS.
Authors’ Contribution
Conceptualization: Thuy T.T. Nguyen (Lead), Chi T.Q. Nguyen (Equal), Tuan H. Le (Equal), Hung V. Nguyen (Equal). Writing – original draft: Thuy T.T. Nguyen (Lead), Chi T.Q. Nguyen (Equal), Tuan H. Le (Equal), Hung V. Nguyen (Equal). Writing – review & editing: Thuy T.T. Nguyen (Lead), Hung V. Nguyen (Equal), Heidrun I. Wergeland (Equal). Supervision: Heidrun I. Wergeland (Lead).
Competing of Interests – COPE
There are no conflicts of interest, and no commercial interests or companies are involved.
Ethical Conduct Approval – IACUC
Every effort was made to minimize animal suffering in this study. The experiments were conducted according to the procedures used at the University of Bergen, Norway, and the regulations for fish experiments at the Governmental aquaculture institutes in Vietnam.
Informed Consent Statement
All authors and institutions have confirmed this manuscript for publication.
Data Availability Statement
All are available upon reasonable request.