Introduction
The heat shock proteins (Hsps) are a highly homologous protein family which act as molecular chaperones to regulate the balance of cell life and death.1 They were produced in response to thermal stress, bacterial and viral infections, UV irradiation exposure, oxygen radicals, heavy metal pollution, or other toxic exposure.2 By assisting in folding nonfunctional proteins to reach functional conformation, Hsps have multiple functions.3
Under adverse conditions such as pathogen invasion, Hsp70 can improve cell resistance by regulating the protein folding modification.4 The human Hsp70 can activate innate immunity by regulating the activity of other proteins.5 Hsp70 could also regulate the adaptive immune response and enhance several cytokine secretions by activating macrophages.6 Recently, it was demonstrated that Hsp70 participates in pathogen infections in shrimp. For example, Hsp70 from Litopenaeus vannamei could be upregulated by white spot syndrome virus (WSSV), and Hsp70 helps shrimp survive WSSV infection by modulating the proPO system and apoptosis.7 Upon infection, hypodermal and hematopoietic necrosis virus (IHHNV) infection, the expression of Hsp70 protein was induced.8 Induction of Hsp70 could enhance tolerance to Vibrio parahaemolyticus infection by activating the proPO system when shrimp was facing non-lethal heat shock.9 Transcription of L. vannamei Hsp70 could be upregulated by the Gram-positive bacterium Staphylococcus aureus and the Gram-negative bacterium Vibrio alginolyticus infection.10 The expression of Fenneropenaeus chinensis Hsp70 in hepatopancreas and gills was significantly induced by heat shock and Vibrio anguillavium challenge.11 Further studies showed that recombinant protein Hsp70 could improve the survival rate of L.vannamei under V. parahaemolyticus infection.9,12
The red swamp crayfish, Procambarus clarkii, is a freshwater crustacean invertebrate native to northern Mexico and the southern United States. P. clarkii is the sixth-largest freshwater aquaculture species in China. In 2021, the output of P. Clarkii in China reached 2,633,600 tons. However, due to increased breeding density and the frequent outbreak of shrimp disease caused by pathogenic bacteria, the crayfish breeding industry has suffered severe losses.13 P. clarkii lacks an acquired immunity and can only rely on innate immune responses to resist the invasion of pathogenic microorganisms.14 Thoroughly analyzing the antiviral defense mechanism and exploring the vital immune factors in P. clarkii is essential in formulating prevention and control strategies of shrimp disease. Hsp70 acts as a “guardian,” which can improve resistance and protect stress when organisms are subjected to environmental stress. Hence, understanding the function and mechanism of Hsp70 in crayfish benefits healthy shrimp culture. In this study, the Hsp70 was identified in P. clarkii, and the spatial distribution and temporal expression of Hsp70 were analyzed after Gram-positive bacteria, Gram-negative bacteria, and virus stimulation.
Materials and Methods
Experimental animals
Healthy red swamp crayfish (P. Clarkii, 10±2 g per crayfish) were collected from an aquaculture base in Changde, Hunan Province, China. Shrimp were cultured in tanks for 5 days at approximately 25 °C.
cDNA cloning and analysis
The total RNA of crayfish was extracted using a TRIZOL reagent. The cDNA was synthesized by the first strand synthesis kit (Simgen, Wuhan, China). The transcript of PcHsp70 ORF obtained from the transcriptome database of P. Clarkii was amplified using the PcHsp70-F/R primers (Table 1). The PCR product was further verified by electrophoresis, and the desired sequence was then purified and ligated into the pUCm-T Vector (Sangon Biotech, China). The product was transformed into DH5α cells, and the positive clones were picked for sequencing (Sangon Biotech, China). Sequence features, multiple sequence alignment, and phylogenic analysis were analyzed as described in a previous study.15 The relevant GenBank registration numbers of aligned Hsp70 are as follows: Cherax quadricarinatus ADM94257.1; Metapenaeus ensis AHY94555.1; Eriocheir sinensis ACF98297.1; Penaeus vannamei XP_027225206.1; Scylla serrata AFI61333.1; Euphausia crystallorophias AIR72267.1; Apis cerana UYC36093.1; Oncorhynchus kisutch XP_020358882.1; Danio rerio NP_001186941.1; Mus musculus AAA37869.1; Salmo trutta XP_029629136.1; Paralichthys olivaceus XP_019941072.1; Homo sapiens BAD96505.1; Bos indicus QGW08891.1; Penaeus monodon AIE88312.1; Penaeus japonicas XP 042878451.1; Pachygrapsus marmoratus CAL68994.1; Portunus trituberculatus ACL37319.1; Cotesia vestalis AGF34718.1; Polyrhachis vicina AGF33487.1; Pteria penguin ABJ97377.1;
Spatial expression analysis
Total RNA from muscle, gill, intestine, hemocytes, lymphoid organ, eye, heart, hepatopancreas, stomach, and brain of three individuals was isolated. Quantitative real-time PCR (qRT-PCR) was executed to detect the expression profile of PcHsp70 mRNA in different tissues. The primers of PcHsp70 used for qRT-PCR are shown in Table 1. The qRT-PCR was performed with a total volume of 20 μL, including 10 μL of 2×MonAmp™ ChemoHS Specificity Plus qPCR Mix (Monad, Suzhou, China), 0.4 μL of each primer, 1 μL of the template, and 8.2 μL of Nuclease-Free water. The qRT-PCR program was performed as described in a previous study.15 The relative expression level of PcHsp70 was calculated according to the 2−ΔΔCT method.16
Immune challenge assays
The mRNA expression profile of PcHsp70 was analyzed after different immune challenges. P. Clarkii (10±2 g per crayfish) was divided into four groups. For the bacterial challenge, 100 μl of V. parahaemolyticus (1×107 cells per shrimp) and 100 μl of Staphylococcus aureus (1×107 cells per shrimp) were used to introduce a challenge of Gram-positive or Gram-negative bacteria, respectively.17 For the viral challenge, 100 μl of WSSV (1×106 copies/mL) was used to introduce the virus challenge as previously described methods.18 For the control group, 100 μl of PBS was injected into shrimp.19 The Hepatopancreas and gills of three individuals were randomly collected to detect the expression of PcHsp70 mRNA at different times after injection.
Results
Bioinformatics analysis of PcHsp70
The ORF of PcHsp70 was 1917, encoding a 638 aa protein. The putative MW and pI of PcHsp70 were calculated as 69.88 kDa and 5.23, respectively. The sequence is predicted to contain phosphorylation sites of 40 serine (Ser), 46 threonine (Thr), and 15 tyrosine (Tyr). 4-598 aa belonged to the Hsp70 domain, and no signal peptide was found (Figure 1). A Dnak characteristic motif and two signature sequences of Hsp70 family proteins were found in the PcHsp70. The Dnak characteristic motif was found in the N-terminal section (IDLGTT-F-S), and the two signature sequences were located in the central part of PcHsp70: IFDLGGGTFDVSIL and IVMVGGSTRIPKQK. Moreover, the PcHsp70 protein also contained an ATP-binding site motif (AEAYLGST), a non-organellar motif (RARFEEM), and a conserved C-terminal sequence (GPTIEEVD) (Figure 1).
Multiple sequence alignment of PcHsp70
Multiple sequence alignment results showed that PcHsp70 showed 95.57%, 91.09%, 90.10%, 88.20%, 87.50%, 87.50%, and 87.26% similarity with C. quadricarinatus Hsp70, Penaeus japonicas Hsp70, Scylla serrata Hsp70, Danio rerio Hsp70, Mus musculus Hsp70, Homo sapiens Hsp70, Apis cerana Hsp70, respectively (Figure 2). The results indicated that the evolution of Hsp70 in P. Clarkii was conserved.
Phylogenetic analysis of PcHsp70
The phylogenetic tree results showed that PcHsp70 belonged to the invertebrate branch and was most closely related to C. quadricarinatus Hsp70, with which it formed a clade (Figure 3).
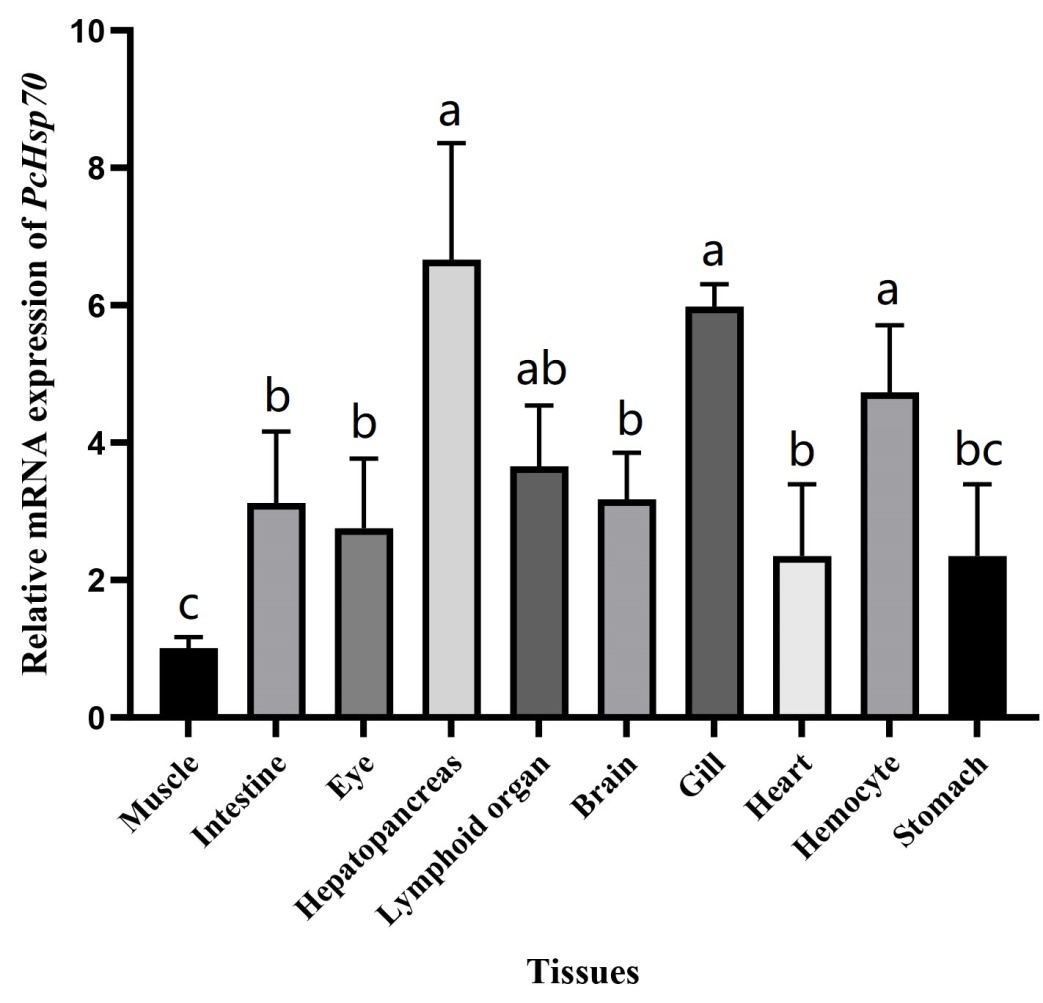
Tissue expression of PcHsp70
qRT-PCR analysis revealed that PcHsp70 was ubiquitously expressed in the ten tissues examined herein. PcHsp70 had the highest relative expression in hepatopancreas tissues, followed by the gill, hemocytes, lymphoid organ, brain, intestine, eye, heart, and stomach. The relative expression of PcHSP70 in muscle was the lowest (Figure 4).
Temporal expression of PcHsp70 post-immune challenge
To further study the immune role of crayfish PcHsp70 during pathogen infection, Expression of PcHsp70 were detected after different immune challenge. When challenged with Gram-positive bacteria (S. aureus), the expression of PcHsp70 in hepatopancreas increased at 6 h post-injection (hpi), peaked at 24 hpi, and then returned to the average level at 48 hpi (Figure 5A). In gill, the expression of PcHsp70 increased at 24 hpi, peaked at 48 hpi, and then returned to the average level at 72 hpi (Figure 5B). Following injection with V. parahaemolyticus, the expression trend of PcHsp70 in hepatopancreas and gill was similar, significantly increasing during 6-48 hpi (Figure 5C and 5D, P <0.01). And the expression of PcHsp70 in the hepatopancreas and gill peaked at 12 hpi (Figure 4C and 4D). After the WSSV challenge, the expression of PcHsp70 in hepatopancreas was increased significantly during 12-48 hpi, while the expression in gill was significantly increased during 6- 48 hpi (Figure 5C and 5D, P <0.05). However, the peak expression in hepatopancreas and gill was 12 phis and 6 hpi, respectively (Figure 5C and 5D).
Discussion
Hsp70 was one of the most well-studied Hsps, which played an essential biological role in improving cell tolerance to stress and maintaining cell self-stability.20 We cloned an Hsp70 from P. Clarkii (PcHsp70) in this study. The PcHsp70 protein contained a Dnak characteristic motif, an ATP-binding site motif, a non-organellar motif, and two signature sequences of the Hsp70 family, indicating that the PcHsp70 had ATPase activity and belonged to cytoplasmic Hsp70 of Dnak type.21 There is a highly conserved cytoplasmic specific regulatory motif at the C-terminal of PcHsp70, which existed in most eukaryotes of Hsp70. This motif affected the activity of the ATPase domain and the binding activity to the substrate and regulated the amount of Hsp70 mRNA transcription in the heat shock state.22,23 Multiple sequence alignment results showed that PcHsp70 had high similarity with other Hsp species in the N terminal ATPase and substrate peptide binding domains. In contrast, its C-terminal domain was less conserved, consistent with the conserved and structural characteristics of the Hsp70 family (date was not shown).24
To preliminarily investigate the role of PcHsp70 in P. Clarkii, the spatial distribution and temporal expression of PcHsp70 were analyzed after different immune stimulation. Tissue distribution results showed that PcHsp70 was widely distributed in different tissues. The highest expression was found in the hepatopancreas, followed by gill and hemocyte. In crustaceans, hepatopancreas was essential for detoxification and immune protection.25 Gills were in direct contact with the external environment and were important immune barriers.26 Hemocytes are essential in immune responses against pathogens, cell encapsulation, and phagocytosis.25 The high expression of PcHsp70 in vital immune organs of hepatopancreas, gill, and hemocyte showed that these tissues were essential sites for PcHsp70 to play the role of immunomodulatory, anti-stress or antioxidant effects. Following S. aureus, V. parahaemolyticus, and WSSV virus stimulation, the expression of PcHsp70 in hepatopancreas and gill was significantly upregulated, suggesting that PcHsp70 might participate in the immune response during Gram-positive bacteria, Gram-negative bacteria, and virus infection in P. Clarkii. There may be several reasons for the increased expression of PcHsp70 in crayfish after being infected with pathogens. In humans, Hsp70 can activate innate immunity by regulating the activity of other proteins.27 In addition, pathogen invasion could induce reactive oxygen species (ROS) or toxic substances, damaging the DNA and proteins.28 As a result of ROS or toxic substances production, the accumulation of denatured proteins might trigger the expression of Hsp70 to solubilize denatured protein, restore protein function, and clear damaged proteins.17 In the present study, a common characteristic of the expression of PcHsp70 in the hepatopancreas and gills after stimulation with S. aureus, V. parahaemolyticus, or WSSV stimulation was that the expression of PcHsp70 was mainly induced between 6-48 hpi, and then tended to be average compared with the control group. This may be due to the production of many ROS or toxic substances in the early stage of pathogen invasion. Hence, PcHsp70 is induced to eliminate the damage of ROS or toxic substances to the host DNA and protein. In the later stages of infection, pathogens may occupy host resources to promote their replication and hence result from inhibiting the expression of PcHsp70. Compared with S. aureus or V. parahaemolyticus stimulation, the expression of PcHsp70 induced by WSSV stimulation was more acute, which increased by up to 4.92-fold at 12 hpi after WSSV stimulation in hepatopancreas. This may be due to the more significant toxicity of WSSV. The regulatory patterns between hepatopancreas and gills are generally similar, but some differences remain. The reasons for the different regulatory patterns between different tissues and between different pathogens need to be further studied. Studies have shown that the up-regulated expression of Hsp70 was beneficial in resisting the invasion of pathogens. In the giant tiger prawn Penaeus monodon, increasing Hsp70 expression could inhibit virus replication and increase shrimp Field’s antiviral ability.29 When the expression of Hsp70 was knocked down by RNAi technology in Artemia franciscana, it reduced tolerance to bacterial infection.30 Therefore, induction of Hsp70 expression after pathogen invasion may be a protective mechanism for crayfish facing environmental stress.
In conclusion, the PcHsp70 was identified in P. Clarkii, and the expression patterns of PcHsp70 were analyzed after different immune challenges for the first time in P. Clarkii. Our results indicated that PcHsp70 was potentially involved in immune responses to Gram-positive, Gram-negative, and virus infections. This will help to understand the significance of PcHsp70 in the immune defense of crayfish. The molecular mechanism and pathway of PcHsp70 involved in immune response need to be further elucidated.
Acknowledgments
This work was supported by the Open Program of Key Laboratory of Cultivation and High-value Utilization of Marine Organisms in Fujian Province (Item number 2021fjscq04); Hunan Provincial Key Laboratory for Molecular Immunity Technology of Aquatic Animal Diseases (Grant No. 2021KJ05); the Education Department of Hunan Province [21C0516 and 21C0515]; the Doctoral Start-up project of Hunan University of Arts and Science [21BSQD11]; Key R&D Project of Hunan Province[2020NK2039]; Hunan Provincial Department of Education Innovation Platform Project[19K064]; Changde City Science and Technology innovation Development Special Fund[2019S044]; and the Innovation Team of Microbial Technology in Hunan University of Arts and Science [202026]; Changde Special Key Project for Scientific and Technological Innovation and Development (No. [2021]67).
Author Contribution
Conceptualization, Writing - original draft preparation: Chao Peng; Resources: Zhongyuan Chen; Writing - review and editing: Qing Han; Software, Methodology: Liye Shao;
Formal analysis and investigation: Ping Mo; Supervision, Funding acquisition: Shuiqing Wu.

_and_deduced_amino_acid_sequence_(below)_of_pchsp70.jpeg)




_and_deduced_amino_acid_sequence_(below)_of_pchsp70.jpeg)