Introduction
Yellow catfish Pelteobagrus fulvidraco is a favorite freshwater aquaculture fish due to its high meat quality, and has been widely farmed in China, Japan, South Korea and Southeast Asia (Li, 2000). Especially in China, the culture of yellow catfish has increased rapidly in recent years with a total production of over 587,800 tons in 2021.1 However, the hemorrhagic disease has become a major bottleneck in yellow catfish aquaculture due to its high mortality.2 Thus, the hemorrhagic disease in yellow catfish should be emphasized to ascertain the sustainable development of this industry. Recently, hemorrhagic disease in cultured yellow catfish has been caused by several bacterial pathogens such as Aeromonas veronii, Aeromonas jandaei, Bacillus cereus, Edwardsiella ictaluri, Shewanella putrefaciens, Staphylococcus sciuri, and Streptococcus iniae,2–8 indicating the diversity of bacterial pathogens from hemorrhagic disease-infected yellow catfish. *Bacillus subtilis *is a facultative anaerobic Gram-positive bacterium that includes virulent members with the ability to produce haemolysins9 and induce high mortality in turbot Scophthalmus maximus and tongue sole Cynoglossus semilaevis.10 Yet scarce information is available on Bacillus subtilis as a causal agent of hemorrhagic disease in cultured yellow catfish.
In this study, a haemolytic isolate HS5 of B. subtilis was demonstrated as a pathogen of hemorrhagic disease in farmed yellow catfish, and its taxonomy, virulence, and antimicrobial susceptibility were further tested. As far as we know, this is the first study to identify haemolytic B. subtilis as a causative agent of hemorrhage disease in cultured yellow catfish. The findings of this study provide insights into the potential threat of haemolytic B. subtilis to cultured yellow catfish.
Materials and Methods
Fish samples
Sixteen hemorrhagic disease-infected yellow catfish (mean weight 3.41±0.51 g) were sampled from infected indoor circular tanks of a fish farm in Fengxian, Shanghai China during November 2022, and were kept in sterile ice-cold bags and immediately transported to the laboratory. Healthy yellow catfish (mean weight 2.11±0.33 g) were acquired from unaffected yellow catfish farming ponds in Huzhou, Zhejiang, China, and were confirmed in good health by sampling a few individuals for careful health examinations according to General Administration of Quality Supervision, Inspection and Quarantine of China.11
Pathogen Isolation
Each sampled diseased fish was externally disinfected with 75% alcohol and dissected according to Yang et al.7 Samples from livers of diseased fish were streaked onto nutrient agar (NA) (Sinopharm Chemical Reagent Co., Ltd., China) with a flamed loop. After incubation for 24h at 280C, isolated bacteria were subcultured on NA plates to check the purity of the isolates. Bacterial suspensions at 5.0×106 colony forming units (CFU)/ml were prepared by suspending bacterial colonies in 50 ml of sterile normal saline,12 which were calculated by counting CFU on NA plates from a series of ten-fold dilutions in sterile normal saline.7 Afterwards, bacterial challenge test was conducted in glass aquaria (80 cm × 54 cm × 50 cm) supplemented with 100 L aerated tap water at 25 0C according to Yang et al.7 Briefly, healthy yellow catfish were randomly assigned to one control and five treatment groups (two replicate aquaria per group, ten fish per aquarium). Treatment groups of fish were injected intramuscularly with 0.1 ml of bacterial suspensions at 5.0×106 CFU/ml, and the control group of fish were injected intramuscularly with 0.1 ml of sterile normal saline. All the test fish were stocked without water change and feeding, and were observed daily for seven days to determine pathological symptoms and mortality.13 The challenge strain was re-isolated from freshly dead fish to confirm the cause of death. Meanwhile, careful examinations of parasites and pathogenic viruses were also carried out as described by Wang et al.14
Pathogen Identification
The genomic DNA was extracted from the pathogenic isolate using a DNA extraction kit (Tiangen Biotech (Beijing) Co., Ltd., China). The near complete 16S rRNA gene was amplified from the genomic DNA by PCR according to Zu et al.15 using primers 27F (5’-AGTTTGATCMTGGCTCAG-3’) and 1492R (5’-GGTTACCTTGTTACGACTT-3’), and the amplified PCR product was further sequenced by Qingdao MDBio Biotech Co. Ltd., China. Following the 16S rRNA gene sequence assembly using Editseq and Seqman in DNAstar software, the alignment of the 16S rRNA gene sequence was performed using the Basic Local Alignment Search Tool (BLAST) program at the National Centre for Biotechnology Information (NCBI), and the phylogenetic tree was further constructed by the neighbour-joining method using the pathogenic isolate’s 16S rRNA gene sequence and its homologous sequences. In addition, the pathogenic isolate was identified phenotypically using API 50CH test strip (BioMerieux, France) as recommended by Zhou et al.16 following the instruction of the manufacturer. The test strip was incubated at 370C and observed after 24h against the API identification index. The phenotypic traits of B. subtilis previously reported by He et al.17 were used as controls.
LD50 Assay
Prior to the virulence assay, the suspensions of the pathogenic isolate at 6.0×105, 6.0×106, 6.0×107, and 6.0×108 CFU/ml were prepared as described above. The virulence assay was performed in glass aquaria (80 cm× 54 cm × 50 cm) supplemented with 100 L aerated tap water at 25 0C according to Yang et al.,7 and consisted of one control and four treatment groups (three replicate aquaria per group, ten fish per aquarium). Treatment groups of fish were injected intramuscularly with 0.1 ml of bacterial suspensions at 6.0×105, 6.0×106, 6.0×107, and 6.0×108 CFU/ml, and the control group of fish were injected intramuscularly with 0.1 ml of sterile normal saline. All the test fish were stocked without water change and feeding, and were observed daily for seven days to determine survival rates.18 Dead fish were immediately sampled to confirm whether the mortality was resulted specifically from the challenge isolate. The mean lethal dose (LD50) value is calculated according to the graphical probit method as recommended by Ogbuagu & Iwuchukwu.19
Virulence Factor Assay
The haemolysis production of the pathogenic isolate was tested with rabbit blood agar (RBA) plates (Guangdong Huankai Microbial Science and Technology Co. Ltd., China), and was observed for the presence of zones around the colony after 24h of growth on RBA plates at 280C.14 In addition, the virulence genes of the pathogenic isolate were also detected by PCR according to Yu et al.20 and Owusu-Kwarteng et al.21 Briefly, the genomic DNA was extracted from the pathogenic isolate using a DNA extraction kit (Tiangen Biotech (Beijing) Co., Ltd., China). The virulent genes encoding enterotoxins (entFM, nheA, nheB, and nheC) were amplified from the genomic DNA by PCR according to Yu et al.20 and Owusu-Kwarteng et al.21 using specific primers listed in Table 1. The PCR products were electrophoresed on 1% (w/v) agarose gel and visualized under ultraviolet illumination. The virulent B. cereus strain CA4, previously isolated from diseased snakehead fish Ophiocephalus argus,14 was used as the control.
Antimicrobial Susceptibility Assay
The antimicrobial susceptibility of the pathogenic isolate was examined in triplicate using the Kirby-Bauer disk diffusion method as recommended by Yao et al.22 Briefly, the suspension of the pathogenic isolate at 1.5×108 CFU/ml was prepared as described above. Afterwards, the pathogenic isolate at 1.5×108 CFU/ml was spread on NA plates, and discs of fourteen antimicrobials (amoxicillin, bacitracin, cefotaxime, cefradine, ceftizoxime, doxycycline, enrofloxacin, gentamicin, florfenicol, kanamycin, netilmicin, norfloxacin, roxithromycin, and sulfamethoxazole) (Hangzhou Binhe Microorganism Reagent Co., Ltd., China) were respectively placed on the inoculated plates. Then, the inoculated plates with antimicrobial discs were incubated at 280C, and the diameters of inhibition zones were measured after 24h of incubation. The antimicrobial susceptibility is determined according to the manufacturer’s instruction.
Results
Pathogen Isolation
A total of five bacterial isolates, temporarily named HS1, HS2, HS3, HS4, and HS5, were isolated from the liver of diseased yellow catfish, and only isolate HS5 was found to be pathogenic to yellow catfish, showing an LD50 of 5.20×106 CFU/ml in yellow catfish (Figure 1). The experimental fish challenged with isolate HS5 displayed the typical hemorrhage sign similar to the naturally-infected fish (Figure 2). The same strain (HS5) that was demonstrated through phenotypic and molecular identification was re-isolated from the experimental diseased fish. In addition, no visible pathological signs or mortality were observed in the control or treatment fish challenged with isolates HS1, HS2, HS3, and HS4, and no parasites or viruses were detected in the naturally diseased fish from which isolate HS5 was obtained. These data indicated that this disease was caused by isolate HS5.
Pathogen Identification
The near complete 16S rRNA gene sequence (1400 nucleotides) of isolate HS5 was submitted to GenBank database with the accession no. QQ926614. A similarity of 99% was noted in the 16S rRNA gene sequence between isolate HS5 and other B. subtilis strains from the GenBank database. The phylogenetic tree further confirmed that isolate HS5 was a B. subtilis strain (Figure 3). Furthermore, the phenotypical features also identified isolate HS5 as B. subtilis, which showed a 100% identity to the reference strain of B. subtilis. It was positive for amidon, amygdalin, arbutin, D-cellobiose, D-fructose, D-glucose, D-maltose, D-mannitol, D-mannose, D-melibiose, D-raffinose, D-ribose, D-saccharose, D-sorbitol, D-trehalose, D-xlyose, esculin, glycerin, glycogen, inositol, inulin, L-arabinose, L-rhamnose, Methyl-αD-glucopyrannosid, N-acetylglucosamin, and salicin, and negative for D-adonitol, D-arabinose, D-arabitol, D-fucose, D-galactose, D-lactose, D-lyxose, D-melezitose, D-tagatose, D-turanose, dulcitol, erythritol, gentiobiose, kaliumgluconat, kalium-2-ketogluconat, kalium-5-ketogluconat, L-arabitol, L-fucose, L-sorbose, L-xylose, methyl-αD-mannopyranosid, methyl-βD-xylo-pyranosid, and xylitol (Table 2). Thus, isolate HS5 was identified molecularly and phenotypically as B. subtilis.
Virulence Factors of Pathogen
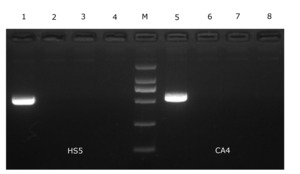
Isolate HS5 formed clear zones of haemolysis on the RBA plate (Figure 4), indicating that isolate HS5 was beta-haemolytic. Furthermore, isolate HS5 was found to harbor the entFM gene (Figure 5).
Antimicrobial Susceptibility of Pathogen
The result (Table 3) showed that isolate HS5 was highly susceptible to enrofloxacin, gentamicin, florfenicol, kanamycin, netilmicin, and norfloxacin, intermediately susceptible to doxycycline, and resistant against amoxicillin, bacitracin, cefotaxime, cefradine, ceftizoxime, roxithromycin, and sulfamethoxazole. These data indicated that isolate HS5 had not developed multiple resistances to quinolones, phenicols, and tetracycline antimicrobials in aquaculture use.
Discussion
Recently, Bacillus species have been documented to cause diseases in aquaculture animals. Isolate LYS1 of B. mycoides has been demonstrated to induce ulcerative disease in largemouth bass Micropterus salmoides.23 Isolate CA4 of B. cereus has been confirmed to cause hepatic hemorrhage of snakehead fish.14 However, there is limited information on B. subtilis as a causative pathogen of yellow catfish. In the present study, we characterized the phenotype, taxonomic position, virulence, and antimicrobial susceptibility of B. subtilis HS5. To our knowledge, this is the first report of haemolytic B. subtilis as a causative agent of hemorrhagic disease in yellow catfish.
The LD50 value is a frequently-used indicator for bacterial virulence assessment.24 B. subtilis strains G7 and NCIB 3610T showed the LD50 values of 3.2 × 105 CFU/g and 4.55 × 105 CFU/g in turbot Scophthalmus maximus, and resulted in hemorrhage signs.10 In our study, isolate HS5 exhibited the LD50 value of 5.20×106 CFU/ml, and also caused hemorrhage symptom which was in agreement with the previous findings.10 As stated by Triyaningsih et al.,25 a bacterial strain can be regarded as virulent when its LD50 value below 107.0 CFU/ml. Thus, isolate HS5 was considered as a virulent strain that could probably cause severe threats to yellow catfish health.
Haemolysin is a primary virulence factor of pathogenic Bacillus species,26 and the haemolysin-producing Bacillus strains have been found to cause bacterial diseases in aquaculture animals.14,27 Several haemolytic strains of B. subtilis have been found to be pathogenic to S. maximus and C. semilaevis.10 In the present study, isolate HS5 of *B. subtilis *was demonstrated to produce beta-haemolysins. This is probably attributed to the involvement of spoVG gene in *B. subtilis *.9 In addition, the virulent entFM gene was also discovered in isolate HS5, which was involve in bacterial adhesion to epithelial cells, vacuolization of macrophages, and virulence.28 The presence of these virulence factors in isolate HS5 probably confer its pathogenicity for yellow catfish.
The antimicrobial resistance in fish-pathogenic Bacillus species has been increasing because of the rising use of antimicrobials to treat fish diseases.29 The largemouth bass-pathogenic B. mycoides LYS1 has been found to develop multiple resistances against cephalosporins, penicillins, and sulfonamides antimicrobials.23 The snakehead fish-pathogenic B. cereus CA4 has been discovered to be multiply resistant to penicillins, polypeptides, and sulfonamides antimicrobials.14 In this study, multiple resistances were also found in B. subtilis HS5 against cephalosporins, penicillins, polypeptides, and sulfonamides. This is consistent with the findings observed in pathogenic B. mycoides LYS1 and B. cereus CA4.14,23 Thus, more attention should be given to the control of fish-pathogenic Bacillus strains. Enrofloxacin and florfenicol are widely used veterinary antibiotics for bacterial disease treatment in aquaculture.30 Dietary supplementation with enrofloxacin at 10 mg/kg body weight for five days has been found to effectively control ulcerative disease in the common carp Cypinus carpio (Liu and Wang, 2004). Oral administration of florfenicol at 5 to 15 mg/kg body weight for ten days has been discovered to significantly control Aeromonas hydrophila infection in gibel carp Carassius auratus gibelio.18 In the present study, B. subtilis HS5 was highly susceptible to enrofloxacin and florfenicol. This serves as a reminder that enrofloxacin and florfenicol can be used to treat B. subtilis infection in yellow catfish. Certainly, some prevention measures should also be taken to reduce the chance of the outbreak of this disease, such as moderate stocking density, less feed waste and routine monitoring of pathogen population.
In conclusion, the findings of this study for the first time identified haemolytic B. subtilis HS5 as a causative pathogen in farmed yellow catfish, and provided new insights into the potential threat of haemolytic B. subtilis to yellow catfish.
Acknowledgements
The work was supported by the Open Fund of Shandong Key Laboratory of Disease Control in Mariculture, the Earmarked Fund for China Agriculture Research System (No. CARS-48), and the Earmarked Fund for Modern Agriculture Fish Industry Technology System of Shandong Province (No. SDAIT-12).
Authors’ Contribution according to CRediT
Methodology: Chunlei Gai (Equal), Xurui Zheng (Equal). Formal Analysis: Chunlei Gai (Equal), Xurui Zheng (Equal), La Xu (Supporting), Jing Diao (Supporting). Investigation: Chunlei Gai (Equal), Xurui Zheng (Equal), La Xu (Supporting), Jing Diao (Supporting). Funding acquisition: Chunlei Gai (Lead). Resources: Xurui Zheng (Lead). Writing – review & editing: Haibin Ye (Equal), Xiaoqing Yu (Equal). Supervision: Haibin Ye (Lead). Conceptualization: Haipeng Cao (Equal), Xiaoqing Yu (Equal). Writing – original draft: Haipeng Cao (Lead).




.png)



.png)
