Introduction
Koi herpesvirus disease (KHVD), resulting from the infection of cyprinid herpesvirus 3 (CyHV-3), has been a global concern for nearly two decades, posing a significant threat to the safety of both carp (Cyprinus carpio) and koi (Cyprinus carpio haematopterus) farming.1,2 CyHV-3, or Koi herpesvirus (KHV), belongs to the Herpesvirales order, Herpesviridae family, and Cyprinivirus genus. It boasts a substantial genome size of 295 kb, encoding 164 ORFs, including 8 duplicate ORFs, making it the largest known herpesvirus in terms of genome size.3
Contemporary research unde-rscores the crucial role of the host’s innate immune system as the primary defense mechanism against pathogenic invasions in fish, often acting more swiftly than the adaptive immune system.4 During the early stages of viral infection, interferons (IFNs) play a pivotal role in initiating the innate immune response against the virus. When the host detects a viral infection through pattern recognition receptors (PRRs), it triggers the innate immune response by recognizing viral nucleic acids and proteins, leading to the upregulation of IFNs.5–7 These IFNs influence viral replication.8 Numerous studies have demonstrated the effective activation of host immune responses during herpesvirus infections.9 Innate immune responses to teleost fish herpesviruses have also been observed, such as Cyprinid herpesvirus-2 (CyHV-2) infection, which induces the expression of immune genes in goldfish.10 Carp challenged by CyHV-3 display robust type I IFN responses.11 However, specific CyHV-3 ORFs’ roles in the infection process remain poorly understood.
In this study, we employed bioinformatics analysis to assess the conservation of ORF24 in cyprinid herpesviruses and conducted colocalization experiments to determine its specific subcellular localization. Our findings revealed that the ORF24 gene of CyHV-3 differs significantly from those in the other two carp herpesviruses, underscoring its unique evolutionary function. Colocalization experiments indicated that ORF24 is distributed in the cytoplasm, and the overexpression of ORF24 effectively inhibited the expression of IFN and its downstream genes stimulated by poly(I:C). These results provide novel insights into CyHV-3-host interactions and lay the groundwork for developing a novel DNA vaccine against CyHV-3.
Materials and methods
Multiple comparisons and evolutionary analyses of amino acid sequences
The study examined the ORF24 gene (GenBank: AVL28280.1) within the CyHV-3 genome (KJ627438) and extracted the amino acid sequence encoding its corresponding protein. A sequence comparison was performed with homologous proteins from three carp herpesviruses, namely CyHV-1 (NC_019491) and CyHV-2 (NC_019495.1) to gain insights into their evolutionary relationships. This comparison process involved using Clustal X 1.83 to align multiple sequences of these homologous proteins. Subsequent searches for similar sequences within the NCBI database were conducted, and conserved domains within these proteins were predicted. The arrangement and presentation of this data were meticulously curated using GenDoc software. MEGA 7.0 software was also employed to construct phylogenetic trees, providing valuable insights into the evolutionary connections among these herpesviruses.
Fish cell lines and plasmids
CCO (Channel catfish ovary) cells and FHM (Fathead minnow muscle cell line) cells were cultured at 25°C in minimum essential medium (MEM) (HyClone, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, New Zealand), penicillin (100 μg ml-1), and streptomycin (100 μg ml-1). Both cells were obtained from our laboratory and stored in liquid nitrogen. The plasmid pEGFP-ORF24 was created by inserting PCR-amplified cDNAs of the ORF24 gene into the pEGFP-N1 vector using the primers listed in Table 1.
Transfection
Poly (I:C) and pEGFP-ORF24 were incubated with TransIntroTM EL Transfection Reagent (TransGen Biotech, China) in 500 μl of Opti-MEM medium (Invitrogen, USA) for 30 min at room temperature. Subsequently, the above reagents were added to the cultured CCO and FHM cells’ culture dishes, respectively. After 6 h of transfection at 25 ℃, the medium was replaced with 1 ml of MEM, and the cells were incubated at 25 ℃. The corresponding cell samples were collected for subsequent RNA sample extraction depending on the experimental conditions.
Quantitative RT-PCR
Total RNAs were extracted from cells using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. The reverse transcription reagent and qRT-PCR reagent used in this experiment were purchased from Vazyme Biotech Co., Ltd. Follow-up experiments were performed according to the manufacturer’s instructions. To detect immune genes, 1 μg of RNA was mixed with 1 μl of Oligo (dT), 4 μl of 4× gDNA wiper Mix, and RNase-free H2O to a total volume of 16 μl. After incubation at 42℃ for 2 min, 4 μl of 5× select qRT supermix II was added and incubated at 50℃ for 15 min and 85℃ for 2 min. The quantitative PCR reactions were conducted in 20 μl volumes, including 10 μl of AceQ qPCR SYBR Green Master mix, 1 μl of cDNA template, 0.4 μl of the forward primer, 0.4 μl of the reverse primer, and 8.2 μl of ddH2O. The cycling conditions were as follows: 95 ℃ for 5 min, 45 cycles at 95 ℃ for 10 s, 60 ℃ for 10 s, and 72 ℃ for 15 s, followed by a final step of 95 ℃ with a 5 °C/s heating rate to create a melt curve. In addition, the reverse transcription experiment used a gradient PCR instrument (Bioer technology, Hangzhou, China); Applied Biosystems QuantStudio 1 Plus (Thermo Fisher Scientific, Suzhou, China) was used in the fluorescence quantitative PCR experiment. Data were normalized to the level of β-actin in each sample using the 2-△△Ct method.12
Statistical analysis
All statistical analyses were conducted using GraphPad Prism 5.0 (GraphPad Software, CA, USA). The statistical significance of the data was assessed using the Student’s t-test, with a significance level set at p<0.05.
Results
Sequence alignment and phylogenetic tree of CyHV-ORF24 and other Cyprinid herpesvirus homology proteins
The bioinformatics analysis of CyHV-3-ORF24 revealed that the protein encoded by CyHV-3-ORF24 comprised 579 amino acids (AA). To gain further insights, we conducted a comparative analysis of its amino acid sequences with those of six homologous proteins from different herpesviruses: CyHV-3-ORF24, CyHV-2-ORF24, CyHV-1-ORF22, CyHV-2-ORF22, CyHV-3-ORF22, and CyHV-3-ORF137. The results indicated that the identity between CyHV-3-ORF24 and CyHV-1-ORF22 was relatively low at 30.75%, and with CyHV-2-ORF24, it was 29.48%. Moreover, the similarity with CyHV-2-ORF22, CyHV-3-ORF22, and CyHV-3-ORF137 was even lower, measuring only 26.52%, 26.51%, and 26.39%, respectively (Figure 1-a). These findings suggest that CyHV3-ORF24 is more closely related to CyHV-1-ORF22 and CyHV-2-ORF24. It also implies that CyHV-3-ORF24 and its homologous proteins may have interactions with the host, potentially contributing to similar diseases caused by this type of herpesvirus. Additionally, when we constructed an evolutionary tree using these homologous proteins, it became evident that CyHV-2-ORF24 was closely related to CyHV-3-ORF24 and distanced from other homologous proteins (Fig. 1-b). This aligns with the results obtained from the amino acid sequence alignment, reinforcing the consistency of our evolutionary analysis.
The Tertiary structure of the ORF24 protein
We employed AlphaFold2 to predict the tertiary structures of CyHV-1-ORF22, CyHV-2-ORF24, and CyHV-3-ORF24. The results indicated that all three proteins can form various secondary structural elements, including α helices, β sheets, and loop structures. Notably, the predominant secondary structure observed in these proteins is the α helix. Specifically, CyHV-1-ORF22 features 18 α helices and 8 β strands, CyHV-2-ORF24 comprises 21 α helices and 9 β strands, and CyHV-3-ORF24 exhibits 22 α helices and 7 β strands. These findings demonstrate that CyHV-3-ORF24 and CyHV-2-ORF24 share a similar tertiary protein structure compared to CyHV-1-ORF22, aligning with our previous sequence alignment results (Fig. 2).
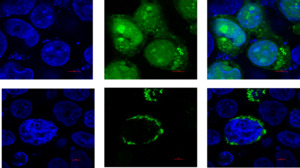
The subcellular localization of ORF24
Upon observing cells transfected with the empty plasmid pEGFP-N1, it became evident that the green fluorescence signal was dispersed uniformly throughout the cells. In contrast, when examining cells transfected with the recombinant plasmid pEGFP-ORF24, the distribution pattern differed significantly from that observed after empty plasmid transfection. In this case, the green fluorescence signal was predominantly localized within the cytoplasm, with some fluorescence observed around the nucleus or dispersed within the cytoplasm (Fig. 3).
ORF24 could inhibit the production of IFN
In this investigation, we focused on determining whether CyHV-3 ORF24 could inhibit the production of type I IFN. As illustrated in Figure 4A-4B, it was evident that CyHV-3 ORF24 significantly suppressed the poly(I:C)-induced IFN production in both CCO cells and FHM cells (Fig. 4). This observation highlights the broad-spectrum anti-IFN activity exhibited by ORF24. Furthermore, we assessed the impact of ORF24 on the mRNA expression of key antiviral genes, namely ISG15, PKR, and TBP, in FHM cells. To do so, FHM cells were co-transfected with either pEGFP-N1 or pEGFP-ORF24 along with poly(I:C). The results unequivocally demonstrated that the overexpression of ORF24 led to a significant reduction in the mRNA levels of ISG15, PKR, and TBP in FHM cells (Fig. 5). These findings strongly suggest that ORF24 possesses the ability to inhibit the expression of antiviral-related genes, potentially promoting the replication of CyHV-3.
Discussion
In this study, we delved into the attributes, localization, and impact on the expression of immune factors of CyHV-3-ORF24. Sequence alignment and a phylogenetic tree analysis revealed that CyHV-3-ORF24 shared a close evolutionary relationship with CyHV-2-ORF24 and stood further apart from other homologous proteins. This suggests the uniqueness of CyHV-3-ORF24 in the context of evolution. Tertiary structure analysis further supported the conservation of CyHV-3-ORF24’s spatial structure, emphasizing its distinct role in the evolutionary process. Simultaneously, we investigated the influence of ORF24 on the mRNA expression of IFN and IFN-stimulated regulated genes (ISRE), specifically ISG15, Mx1, and Viperin. The results clearly indicated that the overexpression of ORF24 significantly diminished the mRNA levels of IFN and ISRE, categorizing it as an immune evasion factor employed by CyHV-3. This study not only reaffirmed the uniqueness of ORF24 in the virus’s evolutionary history but also unveiled its immune evasion capabilities.
Research into CyHV-3 has made significant strides, driven by the urgent need to combat this disease. Currently, vaccines such as DNA vaccines,13,14 inactivated vaccine,15,16 and conventional attenuated vaccine17 represent the most effective means of control. Additionally, diagnostic methods for CyHV-3 have evolved, encompassing techniques like gold nanoparticle-based hybridization assays,18 quantitative polymerase chain reaction (qPCR), conventional PCR (cPCR), virus isolation by cell culture,19 and intra vitam assays.20 The functional roles of ORFs encoded by the CyHV-3 genome have also been elucidated. For instance, ORF134, one of the most abundant proteins of CyHV-3, bears structural similarities to carp IL-10 and promotes the proliferation of IgM1 B cells and memory T cells.21,22 Furthermore, it can inhibit the expression of proinflammatory genes induced by LPS, playing a pivotal role in viral immune evasion.23 ORF57 and ORF131 have both been identified as potential DNA/subunit vaccine candidates,13,24 while ORF4 and ORF12 act as novel viral homologs of the TNF receptor, causing embryonic lethality, morphological defects, and apoptosis.25
As the host’s first line of defense, the innate immune system responds more rapidly than the adaptive immune system. Therefore, we investigated the regulatory mechanisms of CyHV-3 ORFs within innate immunity. Previous research has demonstrated that ORF89 of CyHV-3 functions as a negative regulator of IFN expression by degrading IRF3 and obstructing its nuclear entry through disrupting IRF3 dimerization.26 In this study, we explored the impact of ORF24 on the expression of IFN and its downstream immune genes. The results clearly showed that the overexpression of ORF24 significantly suppressed the expression of IFN and ISRE, although the precise internal mechanisms remain unclear. Notably, RIG-I-like receptors (RLRs) play a pivotal role in activating IFN expression and are conserved in both mammals and fish.27 Additionally, the downstream gene, mitochondrial antiviral signaling protein (MAVS), can associate with the mediator of IFN regulatory factor 3 (IRF3) activation, known as MITA.28 This complex transduces signals to downstream TANK-binding kinase 1 (TBK1), initiating the phosphorylation of IRF3 and IRF7, ultimately leading to IFN expression.29 This cascade then stimulates the expression of IFN-stimulated genes (ISGs). Considering that ORF24 can also inhibit IFN expression, it raises the question of whether ORF24 might affect IFN expression by interacting with these key “IFN regulatory genes”. Previous research had shown the ORF89 of CyHV-3 could interact with the “IFN regulatory genes”–IRF3, and further experiments had also proved that the N-terminal of ORF89 is its main functional domain. Since CyHV-3 is composed of 164 different functional proteins, its specific immune escape mechanism is also relatively complex. Meanwhile, the completion of its escape system cannot be achieved only by the function of a single protein. In this study, ORF24 can also effectively inhibit the increase of IFN stimulated by poly(I:C), showing a certain immunosuppressive effect, but its possible interactions with “IFN regulatory genes” and the underlying molecular mechanisms are unclear and request further exploration (Fig. 6).
In summary, we have summarized the characteristics of CyHV-3-ORF24 and verified its ability to modulate IFN expression, indicating its essential role as a functional protein in virus-host interactions and immune evasion. These findings lay the groundwork for developing future anti-CyHV-3 drugs and disease detection methods.
Declaration of Competing Interest
The authors declared that they have no conflicts of interest to this work.
Acknowledgments
This work was supported by the project of the National Natural Science Foundation of China (32303068) and the Scientific Research Project of Wuhan Customs, Grant/Award Number:2023WK011; The Research and Innovation Initiatives of WHPU (2023)
Authors’ Contribution
Formal Analysis: Jing Wang (Equal), Yan Ji (Equal), Xuan Zhou (Equal). Writing – original draft: Jing Wang (Equal), Yan Ji (Equal), Xuan Zhou (Equal). Resources: Xuan Zhou (Equal), Denghang Yu (Equal), Chi Zhang (Equal). Validation: Denghang Yu (Equal), Kianann Tan (Equal), Chi Zhang (Equal). Supervision: Kianann Tan (Equal), Chi Zhang (Equal). Writing – review & editing: Kianann Tan (Equal), Chi Zhang (Equal).



_cco_or_fhm_cells_were_transfected_with_or.png)
_fhm_cells_were_transfecte.png)




_cco_or_fhm_cells_were_transfected_with_or.png)
_fhm_cells_were_transfecte.png)
