Introduction
Largemouth bass ranavirus (LMBRaV), also called largemouth bass virus (LMBV), is a member of the Ranavirus genus of the Iridoviridae family.1 Seven genera of the family Iridoviridae have been identified: Lymphcystivirus, Megalocytivirus, Ranavirus, Chloriridovirus, Daphniairidovirus, Decapodiridovirus, and Iridovirus. Ranaviruses infect lower vertebrate species including fish, amphibians and reptiles. Furthermore, seven species are explicitly listed in the genus Ranavirus, including Ambystoma tigrinum virus (ATV), Common midwife toad virus (CMTV), Epizootic haematopoietic necrosis virus (EHNV), European North Atlantic ranavirus (ENARV), Frog virus 3 (FV3), Santee-Cooper ranavirus (SCRV), and Singapore grouper iridovirus (SGIV); of these, LMBV belongs to the SCRV. In 1991, an iridovirus was isolated from diseased largemouth bass (Micropterus salmoides) in Lake Weir, Florida, USA, identified as LMBV in 2002.1 The virus was identified in more than 20 states across the USA. In China, LMBRaV was first discovered in Guangdong province in 2008, then in Zhejiang province in 2018.2 In 2021, we identified and isolated a strain of LMBRaV in Hubei province, which named LMBRaV-HB001. The diseased largemouth bass exhibited clinical symptoms such as surface body ulcerations and a swollen and white liver.2 Jiang et al. created a digital PCR assay for LMBRaV that could detect DNA samples with as low as 2 copies/μl (Jiang et al., 2023). However, there is a lack of research on virus morphogenesis and infection mechanism of LMBRaV.
The studies of virus morphogenesis is a critical step to invest the virus life cycle and pathogenesis.3 The host antiviral effects may impose limitations on various stages of the virus lifecycle, such as binding, entry, uncoating, synthesis, transport, assembly and budding.4 Each of these steps is an important target for antiviral prophylaxis and therapy. Ma et al. observed the process of Chinese giant salamander iridovirus (GSIV) entry into cells and morphological assembly using electron microscopy.5 Qin et al. not only studied the capsid assembly of SGIV but also revealed the interaction between virus and endoplasmic reticulum.6 All this research provided valuable data for subsequent studies on antiviral mechanisms. Therefore, we investigated the morphogenesis of LMBRaV in EPC cell line which is susceptible to LMBRaV infection, and the ultrastructural changes caused by virus infections were also analyzed.
Materials and Methods
Cell and virus
The LMBRaV-HB001 was identified in 2021 and propagated in epithelioma papulosum cyprinid (EPC) cells.2 The EPC cell line was obtained from the China Typical Culture Collection Center, Wuhan University. EPC cells were grown in medium 199 (Hyclone) containing 10% fetal bovine serum (FBS) (Gibco) at 25°C.
Viral proliferation curve
EPC cells were plated into twenty-seven T25 flasks one day before infection. Three flasks were reserved as uninoculated controls, while the remaining twenty-four flasks were inoculated with the virus at a multiplicity of infection (MOI) = 0.1 TCID50/cell.7 The cells and supernatants were collected at 2 h, 6 h, 12 h, 24 h, 48 h, 72 h, 96 h, and 120 h post infection (hpi). The cell debris was removed by being frozen-thawed three times and then centrifuged at 3,000 ⅹ g for 10 minutes at 4°C. EPC cells were plated into 96-well plates (Corning, USA) in 10% medium 199 at 25°C overnight. The number of cells per well was approximately 1×104 - 2×104. The virus was diluted with serum-free medium 199 in a 10-fold gradient and then transferred 0.1 mL of each dilution to the 96-well plates. After incubation at 28°C for 1 hour, added 0.1 mL 2% medium 199 in each well. Ten wells were inoculated at each dilution, and two wells without virus were included as control. Cytopathic effect (CPE) was checked daily, and the number of wells with CPE was recorded after 7 days. TCID50 was calculated using the Reed-Muench method. These experiments were repeated three times. The viral titers were log10 transformed, and three replicates were presented as mean ± standard deviation.
Observation of ultrastructural morphogenesis of viruses
The other eight flasks containing monolayer EPC cells were inoculated with LMBRaV (MOI=0.1). After incubation for 2 hours, the inoculum was removed, and 5 ml 2% medium 199 was added. The cells were then incubated at 28°C. The cells were collected at 2 h, 6 h, 12 h, 24 h, 36 h, 72 h, and 96 h post infection. The collected cells were fixed in 2% glutaraldehyde overnight at 4°C.
Electron microscope sample preparation and observation
The fixed cells were post-fixed with osmium tetroxide, then dehydrated using alcohol and acetone, and finally embedded in Epon 812 (SPI). After the ultra-thin sectioning (Leica EM UC7), the sections (50-60 nm in thickness) were stained with uranyl acetate-lead citrate and then examined using a transmission electron microscope (Hitachi H-7650).5
Results
Viral proliferation curve
The dynamics of LMBRaV proliferation in EPC cells indicate that virus-infected cells entered the logarithmic growth phase after 12 hours, achieved the highest virus titer at 72 hours, and then entered the plateau phase (Figure 1).
Virus entry into cells
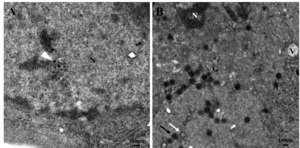
After 2 hours of LMBRaV virus inoculation, it was observed that the virus entered the cell membrane through two methods: (1) direct penetration: virus particles closely approached the cell (Figure 2 A), adhered to the cell membrane surface (Figure 2 B), then virus capsid and cell membrane fused (Figure 2 C) and finally virus particles partly enter the cell (Figure 2 D). (2) endocytosis: the cell membrane formed a pit, and the virus was enveloped in the pit to enter the cell (Figure 2 E-F).
Virus transport in the cytoplasm
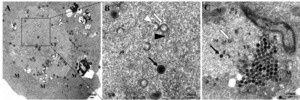
At 4-12 hpi, viral particles mainly existed in three patterns in the cytoplasm: (1) random existing in the cytoplasm (Figure 3 A), (2) enclosed in vesicles (Figure 3 B), (3) entering lysosomes (Figure 3 C). Near the nuclear membrane, the released viral core crossed through the nuclear membrane and entered the cell nucleus (Figure 3 B, D). Autophagic bodies were also indicated in the cytoplasm (Figure 3 B), and mitochondria and the Golgi apparatus were swelling (Figure 3 A-C).
Intracellular replication of the virus
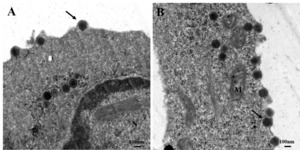
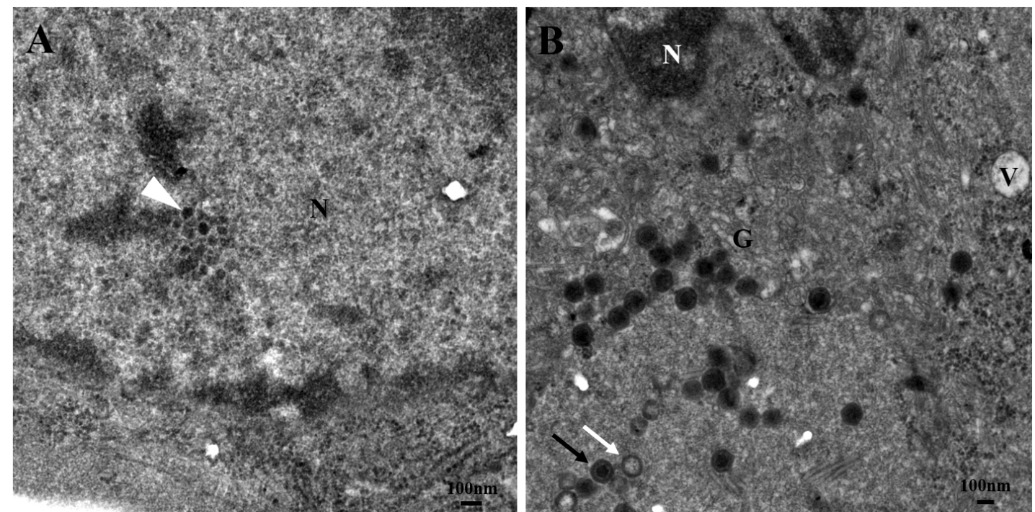
At 24 hpi, virus cores replication was presented in the nucleus, and they were tightly arranged (Figure 4A). At 48 hours, assembling viruses with mature virus particles and empty capsids could be seen in the cytoplasm and cytoplasm vacuoles. Meanwhile, the Golgi apparatus swelled, and the nuclear material condensed (Figure 4B). At 72 hours, a viral factory was visible in the cytoplasm, with intermediate assemblies, empty capsids, mature virus particles, and tubular structures. Around the virus factory, numerous mitochondria became swollen. The vacuoles in the cytoplasm and condensation of nuclear material also occurred (Figure 5A, B). At 96 hours, large numbers of assembled virus particles accumulated around the viral factory and lined up, then the virus particles were arranged in pseudocrystalline array (Figure 5C). The nuclear margination was also observed (Figure 5C).
Release of the virus
After assembly, the virus was released into the extracellular space through budding, the virus particles were wrapped by cell membrane (Figure 6A, B). The nuclear material condensed; moreover, fibrotic nuclear material was observed in the nucleus of the infected cells (Figure 6A).
Discussion
LMBRaV belongs to the Ranavirus genus in the Iridoviridae family. Ranavirus has a wide host range, including amphibians, fish, and reptiles. While amphibian ranavirus, such as FV3 and GSIV, had shown morphological changes of cells post infecton.5,8 LMBRaV, the ranavirus infection with fish, showed genetic sequence differences from amphibian ranavirus.9,10 Recently, the virus genome analysis showed that LMBRaV constituted a unique type in ranaviruses, which was divided from other fish ranaviruses (such as SGIV and EHNV) and amphibian ranaviruses (such as RGV, FV3 and CMTV).11 Although the ultrastructural changes of SGIV have been identified,6 it is still unclear whether there are differences of ultrastructural changes between fish ranavirus and amphibians ranavirus.
To better observe the ultrastructural changes caused by virus infection of host cells, it is crucial to select virus-sensitive cells and determine the optimal time for sample preparation.5 Previously research has indicated that EPC cells were sensitive to LMBRaV.2 Therefore, in this study, we utilized the LMBRaV-HB001 strain isolated in our laboratory to infect EPC cells. Cells were harvested at various time points to create a virus proliferation curve, revealing the infection and proliferation properties of the virus. During 24-72 hours, in the logarithmic growth phase, massive virus replication was also observed within the cells under TEM (Figures 4, 5). After 96 hours, the virus titer ceased to increase, which matched the abundant release of mature virus particles after replication and assembly (Figures 6). These results served as valuable reference information for harvesting high titer virus and creating LMBRaV cell culture inactivated vaccines.
The analysis of virus morphogenetic stages plays a crucial role in studying the interaction between hosts and viruses and in developing virus treatment methods. LMBRaV infection of cells was divided into four stages: adsorption, assembly, maturation, and release. After the virus had adsorbed onto the cell, it entered the cell through direct fusion with the cell membrane or by endocytosis (Figure 2).The infection of FV3 and GSIV utilized the fusion of the virus with the cell membrane and endocytosis, while RGV and tiger frog virus (TFV) entered the cell through endocytosis.5,6,12,13 SGIV entered cells via micropinocytosis and clathrin-medicated endocytosis,14 but no micropinocytosis was detected in this study. This phenomenon might be due to the different virus types. Overall, the methods of LMBRaV entering cells were more similar to that of amphibian ranavirus, which might provide the other evidence that LMBRaV constitute a unique type in ranaviruses as previously reported.11 The process of virus entry into cells is complex, direct fusion with the cell membrane and endocytosis maybe not the only two entry methods of LMBRaV, more research should be performed to detect whether there are more entry patterns.
After entering the cell, LMBRaV shed the capsid in the cytoplasm, allowing the virus core to enter the nucleus for replication. Similar observations have been showed in the morphological changes of amphibian ranaviruse, such as RGV and GSIV.5,15 In this study, it was detected that the virus was present in both the lysosomes and vesicles of the cells. Additionally, it was observed that the virus near the nucleus had shed the capsid, leaving only the viral core (Figure 3). Yang found that lysosomes could degrade viral structures as a mechanism of resisting viral infections, but viruses could also exploit lysosomal disassembly of the capsid to promote viral infection.16 Consequently, LMBRaV might utilize lysosomal disassembly of the capsid to facilitate the release of viral nucleic acid.
Virus has developed complex mechanisms to manipulate host cells as potential ecological niches for their persistence and proliferation.17 This primarily involves binding to host cell membranes and altering specific organelles’ regular structural organization and cellular function.18 The viral components are concentrated in the viral matrix, thus increasing the efficiency of viral replication.6,19 The low electron density of the viral matrix is due to the absence of cell structures. As a virus factory (VF), the viral matrix contains abundant viral nucleic acids and proteins.20 The formation of VFs typically involves rearranging the host cell membrane, reorganizing the cytoskeleton, and recruiting specific organelles such as mitochondria.21 VFs have been reported in studies of several virus replication such as grouper iridovirus (GIV), RGV and herpes simplex virus (HSV).6,22,23 Qin et al. implied that RGV utilized tubular membranes to produce empty capsids that were linked with the creation of fully formed nucleocapsids.6 This study observed low electron density VFs in the cytoplasm at 72 hpi. Tubular structures, assembly intermediates, empty capsids, and mature virus particles were observed in the VFs. There were swollen mitochondria and Golgi apparatus surrounding the VFs, along with vacuoles in the cytoplasm (Figure 5). Therefore, LMBRaV might utilize the organelles and the cytoskeleton to obtain energy and raw materials for virus replication. Once assembly was complete, mature LMBRaV virus particles were budding released outside the cell (Figure 6). Similarly, amphibian ranavirus and other fish ranavirus, such as RGV, GSIV and SGIV, were also released via budding from the cell membranes when assembly had finished.5,6,15 Therefore, budding release might be the main mechanism for the release of ranavirus, including amphibian and fish ranaviruses.
Autophagy is a highly conserved, lysosome-dependent, intracellular catabolic mechanism that removes redundant or damaged organelles and proteins. Therefore, autophagy maintains the health and functionality of the cell.24 It also plays important roles in viral infections. HSV-1 induced the formation of autophagosomes in the infected macrophage, and the virus was engulfed in the autophagosomes and processed by the proteasome. Then the autophagosome virus antigens were presented to MHC class I molecules. In snakehead vesiculovirus (SHVV) infected cells, double membrane autophagosomes were observed in the cytoplasm. Moreover, viral replication could be inhibited by autophagy.25 Qi et al. determined that LMBRaV triggered antiviral autophagic effects, which reduced the viral titer.26 Deng et al. discovered that LMBRaV infection induced numerous autophagosome-like membranous vesicles, and the rapamycin-induced autophagy impeded viral replication and apoptosis.27 In the present study, autophagosomes were observed in cells following LMBRaV infection, which confirmed that LMBRaV-induced autophagy might be a defense mechanism of the cells.
Conclusion
In general, we have revealed basic understandings of the morphogenesis of LMBRaV through analyzing the viral proliferation curve and the ultrastructure observation of LMBRaV replication at different stages. LMBRaV morphogenesis was divided into four stages: (1) adsorption stage: virus adsorbed onto the cell and entered, shed the capsid in the cytoplasm and then the virus core entered the nucleus; (2) assembly stage: virus core replicated, virus capsid formed and mature virus particles assembled; (3) maturation stage: virus factory formed, matured virus particles accumulated and lined up; (4) release stage: matured virus released into the extracellular space. These results contribute to identifying and classifying viral pathogens and promote the development of effective medicine and vaccine to control the LMBRaV disease.
Acknowledgments
This work was supported by the Natural Science Foundation of Hubei Province (2021CFB486), Central Public-interest Scientific Institution Basal Research Fund (YFI 202207), National Natural Science Foundation of China (No. 32202980), Technical Innovation Special Project of Hubei Province (2022BBA0054), National Freshwater Aquatic Germplasm Resource Center (FGRC18537), Central Public-interest Scientific Institution Basal Research Fund (2020TD44).
Authors’ Contribution
Methodology: Mengwei Zhang (Equal), Tao Yang (Equal), Yan Meng (Equal), Chen Xu (Equal). Formal Analysis: Mengwei Zhang (Equal), Yiqun Li (Equal), Yan Meng (Equal), Nan Jiang (Equal). Investigation: Mengwei Zhang (Equal), Yiqun Li (Equal), Yan Meng (Equal), Nan Jiang (Equal). Writing – original draft: Mengwei Zhang (Equal), Wenzhi Liu (Equal), Yan Meng (Equal). Resources: Mingyang Xue (Equal), Yan Meng (Equal). Funding acquisition: Yuding Fan (Equal), Yong Zhou (Equal), Nan Jiang (Equal). Supervision: Yuding Fan (Equal), Yong Zhou (Equal). Conceptualization: Yong Zhou (Equal), Nan Jiang (Equal). Writing – review & editing: Nan Jiang (Lead).
Conflict of Interest
The authors declare that they have no conflicts of interest.
Data Availability Statement
The data supporting this study’s findings are available from the corresponding author upon reasonable request.