Introduction
Interferon regulatory factors (IRFs) serve as critical transcription factors that play a certain role in immune cell apoptosis and differentiation, organism defense against pathogens.1,2 IRF family comprise nine members in mammalian. To be specific, IRF1-9 and a total of 11 members are identified in fish IRF family, except for IRF1~9, IRF10, as well as IRF11.3 IRF10 was first identified in birds, and it was later reported only in fish and birds, whereas IRF11 was only identified in fish.4–7
IRF5, member of IRFs, has been reported as a vital transcription factor mediating innate immune response while bridging adaptive immunity. IRF5 has been demonstrated with profound and diverse regulatory roles in the expression of inflammatory factors, macrophage polarization, B lymphocyte activation and proliferation, plasma cell differentiation, as well as antibody secretion.8–12 In innate immune response, IRF5 exerts vital regulatory effects in regulating downstream immune responses of pattern recognition receptor (PPRs) pathway (e.g., Toll like receptor (TLR) 3, TLR4, TLR7, TLR9, nucleotide-binding oligomerization domain 2 (NOD2), as well as retinoic acid inducible gene I (RGI-I)).13–15 Similar to other IRF family members, IRF5 predominantly resides in the cytosol in a latent form as a monomer in unstimulated cells. IRF5 stimulated by viruses, bacteria, or mutant cells are phosphorylated, homo/hetero-dimerized and then translocated from cytoplasm into the nucleus to modulate the production of cytokines, chemokines, and interferons.2,6,14 Extensive research has been conducted on the gene structure and function of IRF5 in Mammalia, Aves, and Fish,2,16 whereas IRF5 in amphibian has been scarcely investigated.
The black-spotted frog, Pelophylax nigromaculatus, belongs to Pelophylax, Ranidae, extensively distributed in east Asia.17 Since the success of rearing, it on an artificial diet, black-spotted frog has been widely farmed in China due to its growing fast, delicious, and nutritious, taking on great economic significance. However, a considerable number of black-spotted frogs in farms in Hunan Province in south-central China have been subjected to an epidemic meningitis-like disease (i.e., “frog torticollis” or “frog cataract”) since 2016.18,19 epidemic meningitis-like disease brings high lethality to frogs in several days, which is one of the greatest threats to the health of frogs and its industry.18–20 Elizabethkingia miricola was identified as the pathogen to the disease.19–21 The black-spotted frog, a type of vertebrate, has evolved powerful immune system, which comprises innate immune system and adaptive immune system.22,23
The investigation of the immune factors involved in the frog resistance to E. miricola infection and their function can provide basic data for the development of novel methods of preventing epidemic meningitis-like disease. To gain more insights into the role of IRF5 in black-spotted frog resistance to E. miricola infection, the PnIRF5 was cloned in this study, and its patterns of tissue expression in response to E. miricola challenge was studied through quantitative real-time PCR (qPCR).
Materials and Methods
Animals and bacterium sources
Healthy cultured black-spotted frogs weighing 35±5 g originated from a commercial aquaculture farm in Changde (Hunan, China). Black-spotted frogs were kept in two 1 m×1 m×0.6 m canvas pool to adapt for two weeks prior to experiment. The respective black-spotted frog was fed 3-5 Tenebrio molitor twice daily at 08:00 a.m. and 16:00 p.m. No mortality was identified prior to experiment, and all selected experimental frogs were euthanized with MS-222(Sigma-Aldrich, Germany) before sampling. E. miricola was isolated and identified from black-spotted frogs infected with torticollis disease in Changde.
Total RNA extraction and cDNA synthesis
Total RNA from each sample (approximately 100 mg) was extracted with the Trizol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer’s instructions. Total RNA purity and concentration were examined with spectrophotometer (Eppendorf Bio Spectrometer® basic, Germany), and the integrity was examined by electrophoresis with 1% agarose gel. Only intact RNA samples with 260/280 nm ratio between 1.8 and 2.0 were employed for cDNA synthesis. cDNA synthesis was carried out using Revert Aid™ First Strand cDNA Synthesis Kit (ThermoFisher, Carlsbad, CA, USA) following the method recommended by the manufacturer.
IRF5 cDNA cloning
Based on the transcriptome sequencing of black-spotted frogs, specific primers of IRF5 were designed (Table 1) with Primer Premier 6.0 and synthetized by Sangon Biotech Co., Ltd. (Wuhan, China). The middle sequence of PnIRF5 was obtained with primer pairs PnIRF5-1F/PnIRF5-1R and PnIRF5-2F/PnIRF5-2R (Table 1). The PCR amplifications were performed with 1 cycle at 95 °C for 5 min, 30 cycles at 95 °C for 30 s, 56 °C for 45s, as well as 72 °C for 60s. Subsequently, the samples were subjected to a final extension step at 72 °C for 10 min and then stored at 16°C.
The 5’-end and 3’-end cDNA fragments of PnIRF5 gene were amplified with 5’-RACE kit (Sangon, Shanghai, China) and 3’-RACE kit (Sangon, Shanghai, China) in accordance with the method recommended by the manufacturer, respectively. The respective amplified PCR product was examined with 1% agarose gel that was stained with GelGreen (Biosharp, Hefei China). Furthermore, the target DNA fragments were purified with FastPure Gel DNA Extraction Mini Kit (Vazyme, Nanjing, China) and then cloned into pUCm-T Vector (Sangon, Shanghai, China) for sequencing at Sangon biotech Co., Ltd (Wuhan, China).
Sequence analysis
Sequence assembly was obtained based on the SeqMan module of DNAStar 7.0.1 software. The sequence homology of PnIRF5 was investigated using the BLAST algorithm on the website of National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast). The amino acid sequences were translated with the Translate tool of Expert Protein Analysis System (https://web.expasy.org/translate/). The conserved domain of PnIRF5 was predicted with Simple Modular Architecture Research Tool (http://smart.embl-heidelberg.de/). Multiple sequences were aligned using Clustal X2.1 and then visualized with GeneDoc 3.2. The 3D structure of PnIRF5 was predicted by Robetta (https://robetta.bakerlab.org/) and visualized by PyMOL 2.5.2. The neighbor-joining (NJ) method was used to build a phylogenetic tree with 1000 bootstrap replicates using MEGA 11 software.
Tissue specific PnIRF5 expression profiles
Spleen, intestine, liver, muscle, brain, kidney, lung, and stomach were taken from three health black-spotted frogs, respectively. The method of RNA extraction and cDNA synthesis followed the above description. The qPCR reaction was performed using the Roche LightCycler480 II (Roche, Basel Switzerland). The putative housekeeping gene 18S was amplified as an internal control for cDNA normalization with the primer set 18S-RTF/18S-RTR (Table 1). IRF5 gene expression was detected using the primer set PnIRF5-RTF/ PnIRF5-RTR (Table 1). The qPCR reaction system comprised 9.2 μL of the cDNA sample that was diluted with 8 times the volume with nuclease-free water, 10 μL of ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing China), and 0.4 μL of the respective gene specific primer (10 μmol/L). The qPCR cycling protocol covered 1 cycle of 95 °C for 10 min, 40 cycles of 95 °C for 10 s, 60 °C for 10 s, as well as 72 °C for 15 s, followed by a dissociation curve analysis to verify the amplification of a single product. The temperature was elevated from 65 to 95 °C with an increment of 0.5 °C per 5 s, and the data were analyzed using the 2-ΔΔCt method. Furthermore, the results were plotted using GraphPad Prism 6.
Expression profiles of PnIRF5 mRNA after E. miricola infection
Activated E. miricola were incubated at 28 °C for 24 h in brain heart infusion (BHI) broth. The cells were cleaned with sterile PBS and then resuspended in PBS to 1 × 108 CFU (colony forming unit)/mL. The respective black-spotted frog was intraperitoneal (i.p.) injection with 200 μL live E. miricola. The room temperature was maintained at 28 ± 1 °C in the experimental process. Black-spotted frog spleen, liver, and kidney samples were collected at 0-, 1-, 2-, 3-, and 4-days post injection. The method of RNA extraction and cDNA synthesis followed the above description. The mRNA transcription levels of PnIRF5 were determined through qPCR in accordance with the previous description.
Statistical analysis
All data were expressed as mean ± SD from separate experiments. All statistical analysis was conducted using SPSS Version 25.0 (SPSS Inc., Chicago, IL, USA). The one-way ANOVA and Duncan’s MRT was employed for between group comparisons. A probability level of p < 0.05 indicated a difference with statistical significance.
Results
Molecular characterization of PnIRF5
The full-length cDNAs of black-spotted frog IRF5 comprised 2090 bp. The PnIRF5 cDNA (GenBank accession no. OP039101) contained a 356 bp 5’-untranslated region (UTR), a 219 bp 3’-UTR, and an ORF of 1515 bp that was translated into a 504 amino acid putative peptide with a predicted molecular mass of 56.9 kDa. No mRNA instability motifs (ATTTA) and polyadenylation signals (AATAAA) were upstream of the polyA tail (Figure S1). The PnIRF5 covered the conserved domains as follows: a DNA-binding domain (DBD, 1-104 aa), a middle region (MR, 108-195 aa), an IRF association domain (IAD, 196-327 aa), a virus activated domain (VAD, 326-479 aa), as well as two nuclear localization signals (NLSs, 5-10 aa, 413-419 aa) (Figure 1, Figure S1).
As indicated by the BLAST analysis results, PnIRF5 matched significantly with IRF5s from other species. The IRF5 from Rana temporaria (GenBank no: XP_018412577.1) shared the maximum identity (93.85%) with PnIRF5. Besides, PnIRF5 exhibited high identity with other amphibians (e.g., 90.10% identity with Nanorana parkeri (GenBank: XP_018412577.1) and 80.79% identity with Engystomops pustulosus (GenBank: KAG8576680.1)). For Six IRF5s from other species using in multiple sequence alignment, PnIRF5 exhibited the maximum overall similarity (61.26%) to the homolog from Mauremys mutica, exhibiting 83.81% similarity in the DBD and 66.09% similarity in the IAD (Table S1).
Phylogenetic tree analysis
To gain insights into the evolutionary relationships, a phylogenetic tree was built using the NJ method with 1000 bootstrap tests based on the multiple alignments of the IRF5s amino acid sequence. In general, there were five groups in the phylogenetic tree with the Mammalia, Aves, Reptilia, Amphibia, and Osteichthyes with a clear genetic separation. In the Amphibia sub-tree, IRF5 from black-spotted frog was the most closely clustered with R. temporaria (Figure 2).
Tertiary model of PnIRF5
The tertiary model of PnIRF5 was built by Robetta using the RoseTTAFold modeling method. The tertiary structure of PnIRF5 comprised three components that were relative independent in space, i.e., DBD, MR and part IAD, part IAD and VAD (Figure 3A). The DBD was composed of a three-stranded antiparallel β sheet and three α helices; it formed a helix-turn-helix domain. Moreover, five conserved tryptophan were all identified on the surface of DBD (Figure 3B, C). Two NLSs were distributed at the N- and C-terminal of PnIRF5 protein (Figure 3A).
Expression levels of PnIR5 in tissues
qPCR was performed to investigate mRNA expression levels of IRF5 in different tissues of black-spotted frog. As depicted in Figure 4, PnIRF5 transcripts were ubiquitously expressed in all the tissues examined (e.g., the spleen, intestine, liver, muscle, brain, kidney, stomach, skin and lung tissues). The levels notably varied among different tissues. The relative expression of PnIRFF5 was significantly higher in kidney, intestine, lung, spleen, and skin than in other tissues (p < 0.05). No significant difference in PnIRF5 mRNA expression was identified in heart, liver, muscle, and stomach (p > 0.05). Indeed, the maximum expression level of PnIRF5 in the kidney was nearly twelvefold of the lowest expression level in the brain (Figure 4).
Expression of PnIRF5 after Challenged with E. miricola
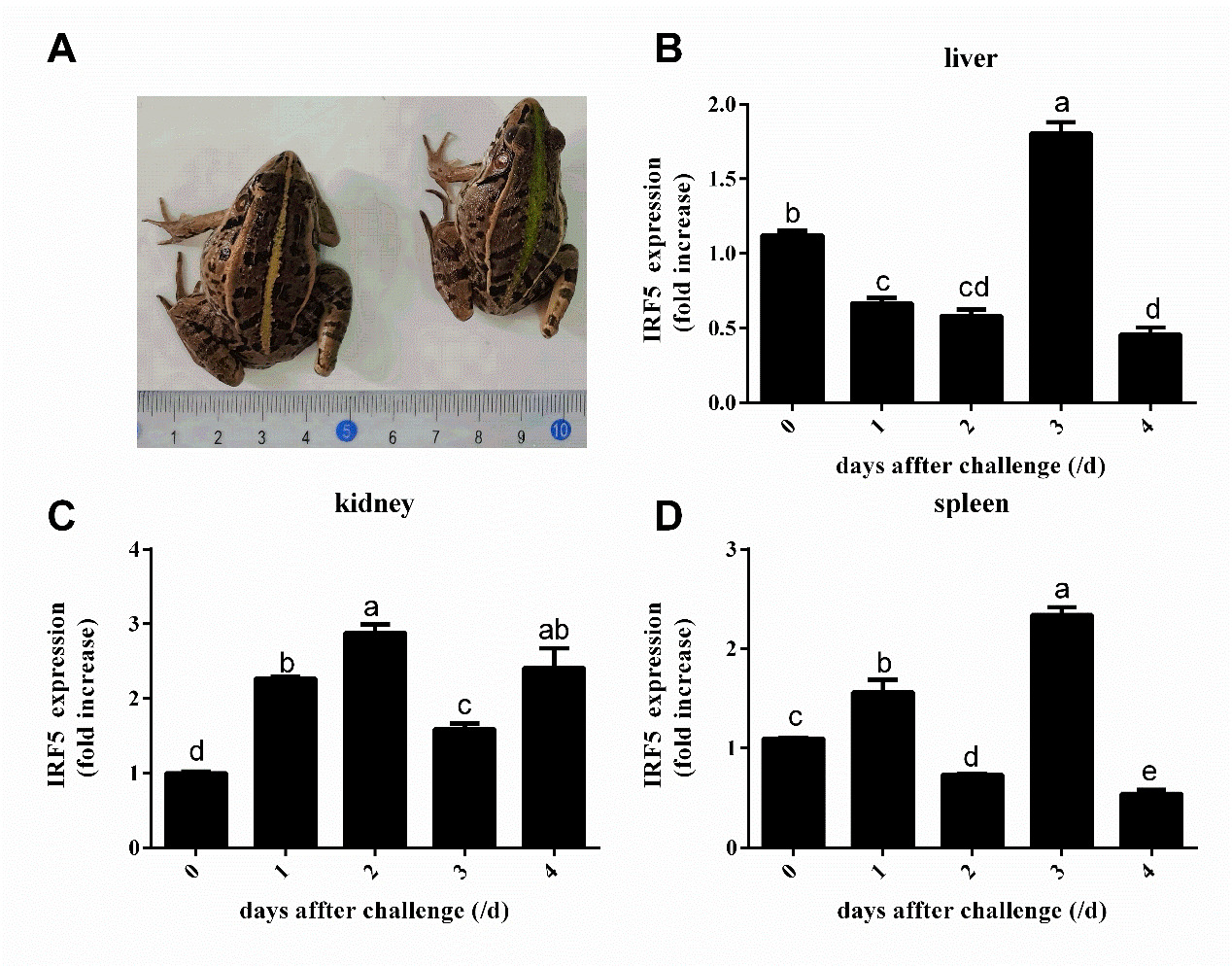
qPCR was performed to detect mRNA expression levels of PnIRF5 in kidney, liver, and spleen tissues at different time points after E. miricola challenge. As depicted in Figure 5, 3 days after E. miricola challenged, a characteristic symptom of slight torticollis was identified in infection black-spotted frogs (Figure 5-A). The mRNA relative expression levels of PnIRF5 in spleen, liver, and kidney displayed significantly up- or down-regulation post E. miricola challenge. For kidney, PnIRF5 expression level was significantly up-regulated at the respective test time point in comparison with 0 days post-challenged. The expression level of PnIRF5 in spleen and liver peaked at 3 days post-injection, and it was peaked at 2 days post-injection in kidney. Furthermore, the maximum expression level of PnIRF5 in liver, spleen, and kidney was 1.80, 2.33 and 2.88 times that at time point 0 days, respectively. In comparison with 0 day post-challenged, the mRNA relative expression levels of PnIRF5 were up-regulated with significance in kidney, liver, and spleen at 3 days after E. miricola challenge (p < 0.05).
Discussion
Vertebrate IRF5 gene has been confirmed as the critical transcription factor for inducing type I interferon. At present, rare studies on IRF5 have been conducted in amphibian. In this study, the IRF5 gene cDNA sequence originated from black spotted frog, and its structure and expression character were explored initially. The full cDNA sequence of PnIRF5 covered an ORF encoding a protein with 504 amino acids exhibiting the typical structural features of the IRF family.
As indicated by the amino acids sequence comparison, the PnIRF5 exhibited an 80.79 - 93.85 % of similarity with other amphibians and a 48.73 - 61.26 % of similarity with other species, suggesting the main structure and function of IRF5 exhibited little diversity between species. PnIRF5 contained a well-conserved N-terminal DBD of 104 amino acids, formed a helix-turn-helix domain, exhibiting five conserved tryptophan repeats, and served as essential domain to bind to IFN-stimulated response element (ISRE, A/GNGAAANNGAAACT) in the promoter region of target genes.24,25 The analyzed tertiary structure suggested that five conserved tryptophan repeats were all on surface of DBD domain. Some previous research has confirmed that tryptophan repeats are a vital component for the protein that can function normally. The W401 residue in human immunodeficiency virus type-1 (HIV-1) reverse transcriptase (RT) Trp-motif takes on critical significance in RT dimerization,26 the W401A mutants were defected for dimerization and devoid of RT activity.27 Zhang28 suggested that the Nanog tryptophan repeat region refers to the location of repression function of Nanog and represents a vital module of Nanog in fine-tuning the balance between self-renewal and differentiation. The contributions of tryptophan repeats in DBD of IRF5 remain unclear, which the elucidation of the immunological regulatory mechanism of IRF will be an interested research topic. The IAD domain of IRF5 in species showed lower conserved in comparison with DBD domain. The lower conserved IAD domain at the C-end results in a wide range of functional diversity of IRFs, which endows IRF family members with the ability to display differential transcriptional activity of different IFN genes or ISGs in different cell types or under different viral infection conditions.6 IAD can mediate interaction with itself, other IRF family members, or other transcription factor family members, and regulate timely and appropriate expression of IFN gene in specific cell classes through synergism or antagonism.24,29 PnIRF5 also contains a VAD at C-terminal, VAD possesses many conserved serine residues, which may serve as virus-induced phosphorylation sites.30 Phylogenetic analysis indicated that IRF5 from black-spotted frog was the most closely clustered with R. temporaria. Collectively, this evidence suggests that PnIRF5 belongs to the IRF5 family member.
The tissue expression characteristics of IRF5 have been investigated most in the fish, and IRF5 was expressed in almost all examined tissues of the fish, whereas the tissue distribution of IRF5 was different in different fish.31–35 In this study, the PnIRF5 was primarily expressed in kidney, intestine, and lung and moderately expressed in spleen and skin, consistent with the fish species (Danio rerio, Mycobacterium tuberculosis).36,37 The possible reason for the differential tissue distribution of IRF5 may be the diverse potential immune systems between Fishes and amphibians. The spleen and kidney serve as crucial immune organs in amphibians, which have a rich resident population of macrophages and lymphocytes. Furthermore, the lung, intestine and skin are vital mucosal immune tissues of amphibians. IRF5 was highly expressed in the above-mentioned tissues, suggesting that PnIRF5 may take on crucial significance in the activity of the immune system in black spotted frog.
E. miricola is highly pathogenic to black spotted frog, and no cure has been reported for the control of its infection. The expression profiling of PnIRF5 in three immune-associated tissues was examined to gain insights into the role of PnIRF5 in response to the infection with E. miricola. After challenged with E. miricola, PnIRF5 expression level all tended to be increased significantly in liver, spleen, and kidney. The similarity results were identified in spleen, head kidney, and liver of Cyprinus carpio L.35 and Cynoglossus semilaevis,34 IRF5 gene was significantly up-regulated upon LPS and Vibrio harveyi challenges, respectively. Furthermore, Pandey et al. suggested that after Mycobacterium tuberculosis infection, type I IFN expression can be stimulated through a pathway that is dependent on NOD2, RIP2, TBK1, and IRF5 in mice.36 Notably, the up-regulated expression of IRF5 after E. miricola infected may be required to play a certain role in the immune response.
Conclusions
In this study, the PnIRF5 genes of Pelophylax nigromaculatus was cloned with a coding sequence length of 1515 bp, encoding 504 amino acids. PnIRF5 genes were expressed in all tested tissues, and the maximum expression was identified in the kidney. Under E. miricola stress, the maximum expression level of PnIRF5 in the liver, spleen, and kidney was 1.80, 2.33 and 2.88 times that at time point 0 days at 3 d, 3 d, and 2 d, respectively, suggesting that PnIRF5 participated in the immune response of P. nigromaculatus resistance to E. miricola infected.
Acknowledgments
This research was funded by the Natural Science Foundation of Hunan Province (Grant no. 2023JJ30435 and 2022NK4150), the Research Project of Education Department of Hunan Province (Grant no. 22B0701 and 22B0691) and the National Natural Science Foundation of China (Grant no. 31972835).
Author contributions
Conceptualization: Ronghua Wang, Pinhong Yang;
Methodology: Ronghua Wang, Ke Li, Menglu Yan, Yan Kang, Shuqiong Li;
Formal analysis and investigation: Ronghua Wang, Qing Tan;
Writing - original draft preparation: Ronghua Wang;
Writing - review, and editing: Ronghua Wang, Pinhong Yang, Zhongyuan Chen;
Funding acquisition: Pinhong Yang;
Resources: Qing Tan, Hongchun Jin, Jinlong Wang;
Supervision: Pinhong Yang.